The building block of life is often heralded as the cornerstone of a new generation of electronic devices and computers, but the electronic properties of DNA remain highly controversial.
DNA plays a pivotal role in biology as the carrier of genetic information in all living species. Recently, however, physicists and chemists have become increasingly interested in the electronic properties of the “molecule of life”. According to some, DNA is a molecular wire that can conduct charge carriers with virtually no resistance. Others, however, find that DNA behaves as an insulator. Experiments are now starting to provide the first clues about the mechanisms that underlie charge transport in DNA.
Despite the current hot debate, the subject is far from new. Soon after James Watson and Francis Crick discovered the double-helix structure of DNA in 1953, Daniel Eley and D I Spivey were the first to suggest that DNA could serve as an electronic conductor. The field has recently been revived with the advent of measurements on single DNA molecules, in particular by Jacqueline Barton’s group at the California Institute of Technology. Barton and colleagues measured the fluorescence produced by an excited molecule and found that it no longer emitted light when attached to a DNA molecule. Their results suggested that this “fluorescence quenching” was due to the charge on the excited donor molecule leaking along the length of the DNA to a nearby acceptor molecule. In other words, they suggested that DNA was a conducting molecular wire.
After much initial controversy, the chemists working on the problem of DNA conductivity are now moving towards a consensus view, as illustrated by the titles of articles in Chemical & Engineering News that have evolved from “DNA: insulator or wire?” in 1997 to “DNA charge migration: no longer an issue” in 2001. It now appears that charge carriers can hop along the DNA over distances of at least a few nanometres. However, the situation among physicists who are measuring electronic transport through DNA molecules over larger distances is much less clear.
Moving electrons from site to site
The process of electron transfer – the movement of charge from one molecule to another, or from one end of a given molecular structure to the other – is one of the most fundamental in chemistry and materials science. Such reactions are crucial in processes ranging from corrosion to photosynthesis. Modern investigations of electron-transfer reactions began in the 1950s with reaction-rate measurements by Henry Taube’s group at Stanford University and others. At the same time, Rudy Marcus at the Polytechnic Institute of Brooklyn and Noel Hush of the University of Sydney began developing theoretical models to describe electron-transfer processes. Their approach combined general ideas of chemical kinetics with specific insights into how the internal motions of molecules can facilitate electronic movement between sites. These mechanistic pictures were later generalized to include quantum-mechanical effects, particularly by Joshua Jortner at Tel Aviv University in Israel and Marcus, and we now have an almost complete understanding of thermal and photoexcited electron transfer.
Consider, for example, an extended molecule with a donor group at one end and an acceptor at the other end. Extensive experimental and theoretical studies have shown that electron-transfer reactions within such a single molecule can occur by two principal mechanisms. The first consists of a single-step electron-tunnelling process from the donor to the acceptor. This process is said to be “coherent” in the sense that the electron does not exchange any energy with the molecule during the transfer, and the electron is never localized. The rate of such reactions decreases exponentially with the distance between donor and acceptor. Therefore, for electron transfer over very long distances, one expects this coherent rate to be insignificant on any reasonable timescale.
The second possible mechanism for long-distance electron transfer is generally referred to as “thermal hopping”. In this incoherent process, the electron is localized on the molecule and exchanges energy with it. Electron transfer proceeds in a multi-step fashion from donor to acceptor. Such hopping processes can transfer charge over far longer distances than the coherent tunnelling process, and the motion can be thought of as diffusive.
Electron transfer within short DNA molecules
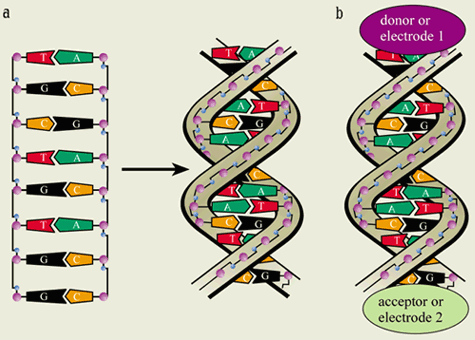
The famous double-helix structure discovered by Watson and Crick consists of two strands of DNA wound around each other (figure 1). Each strand has a long polymer backbone built from repeating sugar molecules and phosphate groups. Each sugar group is attached to one of four “bases”. These four bases – guanine (G), cytosine (C), adenine (A) and thymine (T) – form the genetic alphabet of the DNA, and their order or “sequence” along the molecule constitutes the genetic code. In double-stranded DNA, hydrogen bonds between the bases couple the two strands together. The chemical bonding is such that an A base only ever pairs with a T base, while a G is always paired with a C. The base pairs look like the rungs of a helical ladder. Since the phosphate groups on the backbone are negatively charged, the DNA is usually surrounded by positive “counterions”. The DNA structure is ideal for electron transfer because some of the electron orbitals belonging to the bases overlap quite well with each other along the long axis of the DNA. These so-called stacking interactions also underlie many one-dimensional molecular conductors, including one of the most widely studied organic conductors, TTF-TCNQ.
Early measurements of electron transfer in DNA were performed with a variety of techniques, notably by John Warman and co-workers at Delft University of Technology using microwave conductivity. But the intense interest in charge transfer in DNA began with important work by Barton’s group at Caltech, and by Nick Turro of Columbia University in New York and collaborators. These groups reported fascinating distance dependencies, including the apparent coherent transfer over distances as long as 4 nm. The observations led to the suggestion that DNA acts as a “molecular wire”, and that DNA systems represented a “new paradigm” for electron transfer.
The idea that electron transfer is enhanced in DNA appears quite reasonable. Indeed, the possibilities for testing electron transport in DNA are great because the molecule adopts many different structures. These include kinks, bends, bulges and distortions along the molecule, as well as the “polyelectrolyte” character of the double helix, which may lead to the flow of positively charged counterions along the negatively charged phosphate backbone. A major factor is the sequence of the base pairs along the molecule. It is this extensive chemical variability that leads to various different results observed in DNA electron-transfer experiments. Extensive experimental and theoretical work over the past 15 years has led to substantial clarification of charge-transfer mechanisms in DNA. The dominant mechanisms appear to be the two discussed above – coherent tunnelling and thermal hopping.
From contradiction to consensus
Early experiments yielded apparently contrasting results. This was partly due to the variety of DNA sequences, layouts and conditions. To understand why the DNA sequence makes a difference, we need to compare the relative energies of the G-C and A-T base pairs. These energies have now been deduced from computational models, photoemission experiments and electrochemical measurements.
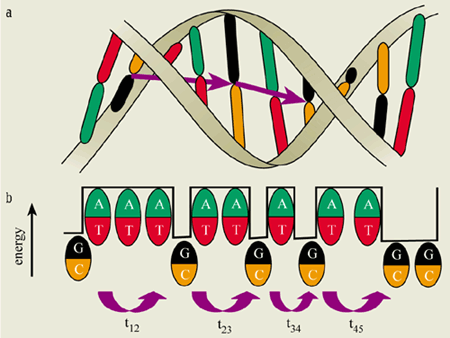
The important feature is that a hole (i.e. a positive charge) is more stable on a G-C base pair than on an A-T base pair (see figure 2). Also, the energy difference between these two pairs is substantially larger than the thermal energy of the charge carrier. Under these conditions, a hole will localize on a particular G-C base pair. Because the A-T base pairs have a higher energy, they act as a barrier to hole transfer. However, the hole can tunnel in a coherent fashion from the first G-C site to the second, and can then either hop back to the first G-C pair or move on to the next one.
The rate of coherent charge transfer decreases exponentially with the distance travelled. But when the distance between G-C base pairs becomes too long for coherent charge carriers to jump efficiently, thermal hopping becomes the dominant charge-transfer mechanism. As an analogy for such motion, consider leaping across a stream. While it is easy to cross a narrow brook in a single jump, it is impossible to leap across a wide river unless there are stepping stones along the way. The overall time it takes to cross the river is then simply the sum of the times it takes to jump from one stepping stone to the next.
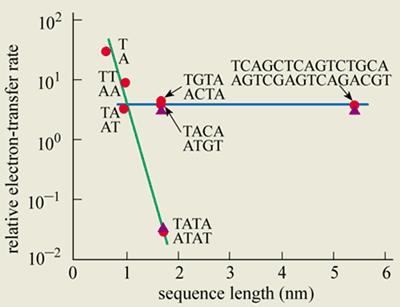
Both charge-tunnelling and thermal-hopping mechanisms have been verified in experiments, notably by Bernd Giese’s group at the University of Basel in Switzerland and by Maibi Michel-Beyerle and co-workers at the Technical University in Munich (figure 3). Meanwhile, George Grüner and co-workers at the University of California at Los Angeles have measured the hopping mobility in DNA and compared it with traditional one-dimensional conductors. And Fred Lewis, Mike Wasielewski and Robert Letsinger at Northwestern University in the US have directly observed both thermal hopping and coherent transfer. Barton and Ahmed Zewail, also at Caltech, have explained their fluorescence-quenching experiments using these two mechanisms.
While this picture of coherent transfer and thermal hopping appears to describe the basics, it may not be the full story. For example, what is the role of local thermal motions of the bases? And do the charge carriers have a “polaron'” character: that is do they distort the neighbouring DNA structure? Such open questions are currently under study.
To sum up, DNA serves as an elegant model for one-dimensional charge transport. The two most fundamental processes for electron transfer in extended electronic systems – coherent tunnelling and diffusive thermal hopping – have been clearly demonstrated in DNA charge-transfer studies. It appears that electrons and holes are indeed able to shuttle along a single DNA molecule over a distance of a few nanometres.
Wiring electronics with DNA?
While recent research within the chemistry community seems to converge on a clear picture for the relevant electron-transport processes at the microscopic level, direct electrical measurements on long DNA molecules by a number of physics groups have yielded conflicting results.
A direct measurement of the resistance of single DNA molecules can be made by hooking the molecule up between two metal electrodes and measuring the electric current running through it. Indeed, measurements of small conducting wires – even those formed from single molecules, such as carbon nanotubes – have emerged in the field of mesoscopic physics over the last few years. Such experiments take advantage of the tools developed for nanotechnology, including electron beam lithography for the fabrication of very tiny nanoelectrodes and atomic force microscopy for imaging samples at the molecular level.
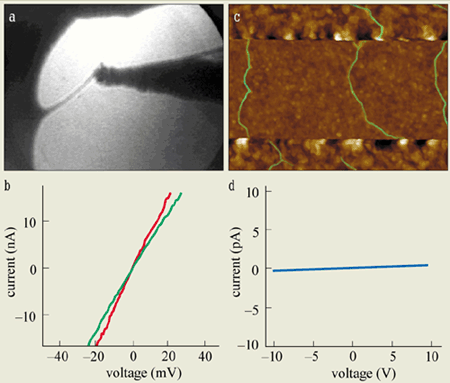
The first direct electrical measurements on small bundles of DNA were made in 1999 by Hans-Werner Fink and Christian Schönenberger at the University of Basel in Switzerland. They developed a special high-vacuum low-energy electron microscope that could image thin free-standing bundles of DNA stretched across a hole in a membrane. They could also measure the conductance (i.e. the inverse of resistance) by touching the DNA bundles with an additional metal tip (figures 4a and b).
The surprising result of this technical tour de force was that DNA bundles almost 1 µm in length appear to behave like an ohmic conductor (i.e. the current rises linearly as the applied voltage is increased). Equally surprising was the low value of the resistance (~1 M ohm), which indicates that DNA conducts well. This was quite unexpected because in the simplest picture one expects that DNA will be a semiconductor with a large energy gap between the valence and conduction bands – in other words DNA is expected to be an insulator.
This finding challenges the current understanding of DNA charge transfer since the molecules used in these experiments had a random sequence of G, A, C and T bases, rather than a large number of G-C pairs. This randomness – together with any additional disorder – should readily localize the carriers and thus prevent the DNA from conducting. The Basel experiments on DNA bundles confirmed earlier experiments by Yoshio Okahata and co-workers at Tokyo Institute of Technology who measured a finite conductivity for stretched films of DNA.
Soon afterwards, Danny Porath and co-workers in one of the author’s (CD) group at Delft made a first attempt at addressing the issue of the DNA sequence using direct-transport experiments. Inspired by the models that emphasize the importance of G-C base pairs for hole transfer through DNA, they made measurements on very short DNA molecules comprising one strand that contained only G bases and a second strand that contained only C bases.
Using a technique called electrostatic trapping, Porath and co-workers placed these molecules between two nanoelectrodes only 8 nm apart. On applying a voltage to the device, they first observed an insulating gap (i.e. no current at low voltage), but above a threshold of about 1 V, they obtained a current through the device. The results suggested that this particular type of DNA – called poly(dG)-poly(dC) DNA – is a large-gap semiconductor (i.e. an insulator). Transport can only occur in these materials when a very large electric field is applied. Such a field may cause the molecular energy bands to align with the energy levels of the electronic carriers in the electrodes, thereby allowing charge carriers to enter the DNA.
More recent experiments by Arnold Storm and co-workers at Delft have measured the electronic conduction at length scales of 40 nm and longer. Convincing images of DNA molecules positioned between nanoelectrodes were obtained with an atomic force microscope, but no conduction was observed for such long lengths of DNA (figures 4c and d). In other words, Storm and colleagues measured an infinite (>1013 ohm) resistance. While these findings contrast with the Basel results, they confirm earlier measurements by Erez Braun and co-workers at the Technion in Israel, who also observed insulating behaviour for DNA some 16 µm in length.
Wide-ranging behaviour
A spectacular report was made earlier this year by Alebker Kazumov, Hélène Bouchiat and co-workers at the Université Paris-Sud. They placed DNA on carbon-covered electrodes that were made of a thin (5 nm) layer of rhenium, a superconducting material. With this set-up they found that room-temperature bundles of DNA have a very low resistance, of the order of 100 k ohm, which approaches the resistance quantum, h/2e2 = 12.9 k ohm, where h is the Planck constant and e is the charge of an electron. (The resistance quantum is the lowest resistance measurable in a perfect metallic wire.) Kazumov and co-workers also reported that DNA supports a “proximity-induced” supercurrent when cooled below 1 K, the temperature at which rhenium becomes a superconductor. This result suggests that DNA behaves as a good conductor and a phase-coherent metal, which is rather puzzling. If the results are confirmed, however, they will be a remarkable finding.
Other experimental results come from the group of Tomoji Kawai at Osaka University in Japan, which has studied DNA extensively using scanning tunnelling microscopy. Recently this group has applied scanning-probe techniques to investigate DNA conduction by measuring the current from the tip of the microscope probe through a DNA bundle connected to a metal strip that acts as a second electrode. Interestingly, they found that the resistance increases exponentially with distance. Indeed, the resistance they measured was high, ranging from 109 ohm to 1012 ohm. They also found that DNA consisting only of C and G bases conducted somewhat better than DNA with A and T bases, which is consistent with the expectations of electron-transfer theory. These interesting findings contrast with results by Pedro de Pablo and co-workers at the Universidad Autónoma de Madrid who found insulating behaviour using the same technique, albeit on “random-sequence” DNA.
DNA can exhibit a surprising range of structural forms and modifications. One interesting example is the replacement of certain hydrogen atoms in the base pairs of the DNA with metal ions. The conduction through such metal-DNA has recently been studied by Andrei Rakitin and co-workers at Brown University in the US, in collaboration with researchers at the University of Saskatchewan in Canada. They reported metal-like current-voltage curves, but they also found that conventional DNA showed low-gap semiconducting behaviour, which again contrasts with measurements by others.
So, where does the field currently stand on the issue of the conduction properties of DNA? It is clear that claims range all over the place: everything from well-insulating behaviour to the support of superconducting currents through DNA has been reported. It is not yet clear whether these opposing results are due to possible artifacts in some experiments, or if they reflect the large “parameter space” of DNA. Possible differences could be due to the base sequence or the length of the DNA, or the properties of the buffer solution in which the DNA is kept. Other factors include the ambient surroundings (whether the experiments are conducted in liquid, air or vacuum), the structural form of the DNA and its organization (whether the bulk material is studied versus bundles or single DNA molecules), and the electrode-molecule interface and so on.
The present authors are unaware of any independent verification of metal-like behaviour and we are confident that long DNA molecules are true insulators, as shown most convincingly by Storm and co-workers. The current status of the field imposes a particularly heavy responsibility on experimentalists. It is more important than ever to devise careful control experiments and for all the findings to be thoroughly reproducible.
Beyond 2001: further along the DNA odyssey
The Watson-Crick double-helix structure for DNA is almost 50 years old. During the last half century, the majority of research into DNA has been devoted to its biological properties, in particular its role in genetic inheritance, disease, aging, RNA synthesis and mutation. Biologists have developed methods to identify and extract sections of DNA and then combine them with other sections with great precision to create new genetic material. Indeed, these same advances in genetic engineering also permit the use of DNA as a component for molecular electronics circuitry.
While it seems unlikely that one can use the intrinsic conductance properties of DNA for single-molecule electronics, Erez Braun, Uri Sivan and co-workers at the Technion have suggested an entirely different approach to DNA electronics by exploiting the molecule’s assembly properties. First they added “sticky ends” to the ends of the DNA – small pieces of single-strand DNA that can bind to “complementary” bases of other sticky ends. Next they attached sticky ends to two metal electrodes so that a single DNA molecule would bridge the gap between the electrodes. The sequence of these single-strand pieces ensured that the binding was highly selective: only those fragments with the matching genetic code coupled to the electrode. After the DNA had assembled, the Technion team replaced the counterions with silver ions so that the DNA could then be used as a seed to grow a thin metallic wire. In this approach, the DNA acts as a linear template for conducting metallic wires.
The highly specific binding between DNA strands may provide a key element in the development of single-molecule electronics. Although many individual components for molecular electronics have been demonstrated recently, a strategy for the assembly of integrated circuits is still largely lacking. The geometry and connectivity of future electronic circuits could perhaps be controlled by DNA self-assembly, rather than by destructive lithography techniques. Indeed, the unique structural properties that control the DNA double helix will almost certainly lead to extensive applications because the synthetic variability and recognition properties of DNA are truly unrivalled by any other molecular structure. One can choose DNA molecules to recognize other DNA molecules, to fill a space in a particular fashion, or to provide a structural scaffolding for the assembly of complex nanostructures.
DNA can be combined with basically any chemical side group imaginable. For example, single-strand sticky ends can be attached to electrically active molecular elements, such as metal clusters, fullerenes or certain molecular switches. These groups can be located at specific sites on the molecules as defined by the sequence of bases. Chad Mirkin, Robert Letsinger and collaborators at Northwestern, as well as Paul Alivisatos’s group at the University of California at Berkeley, have used this idea to prepare nanocomposite structures consisting of gold nanospheres covered with single-strand DNA fragments. They are devising new colloidal materials made from gold spheres joined together with DNA.
The beauty of such materials is that the spacing between the spheres – and thus the thermal, electrical and optical properties – is determined by the nature of the DNA. This spacing can be controlled either statically (by using different sticky ends) or dynamically (by exploiting the melting properties of DNA with changes in temperature or solvent). Such materials can form the basis for remarkably accurate biosensors.
The potential of DNA
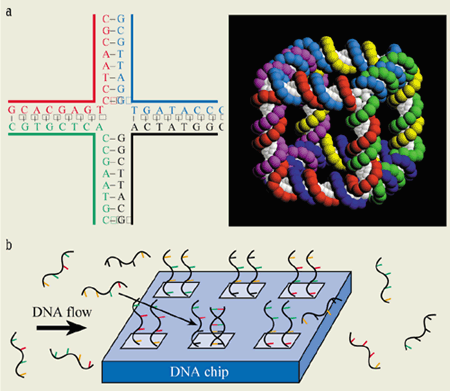
One existing application of the assembly properties of DNA is in so-called DNA chips. These devices exploit the fact that short single strands will bind to other segments of DNA that have complementary sequences, and can thus be used to probe whether certain genetic codes are present in a given specimen of DNA (figure 5).
Microfabricated chips with many parallel DNA probes are becoming widespread in analytical and medical applications. Currently, the chips are read out optically, but further miniaturization might require new read-out schemes, possibly involving the electron-transfer properties of DNA. Such schemes might exploit the different electrochemical responses of single- and double-strand DNA molecules that attach to a surface. However, a detailed understanding of these interfaces remains to be developed.
DNA has the potential for assembling networks with a variety of geometries. Although short pieces of double-strand DNA (below about 50 nm) essentially form a linear stack, intricate spatial structures have been produced by exploiting the self-assembly properties of DNA with sticky ends. In particular, Nadrian Seeman and co-workers at New York University have made intertwined loops, knots, one- and two-dimensional arrays, three-dimensional cubes and nanolattices based on synthetic DNA structures (figure 5). In these cases, the biological properties of DNA are largely irrelevant: the molecule is used simply as a basic building material for nanostructures.
The unique assembly properties of DNA together with its unparalled recognition, optical characteristics, stability and adaptability suggest that DNA may become one of the most important species in the general area of molecular electronics. Often, molecular electronics is viewed only as the formation of logic devices and memories with single conducting molecules. But it can also be defined more broadly as the area of science and technology that studies electronics and sensors based on molecular organization. There is little doubt that DNA is destined to be a major component in the toolkit of molecular electronics.