Physicists in the US have developed a new algorithm to determine the 3D structure of a nanoparticle using 1D X-ray or neutron diffraction data – a feat hitherto impossible. The technique, devised by Simon Billinge, Pavol Juhas and colleagues at Michigan State University, involves compiling a "league table" of possible structures that is constantly updated as the calculation proceeds with good candidate structures being promoted and less good ones being relegated. The approach has been dubbed the "Liga" algorithm after Spain's La Liga football league (Nature 440 655).
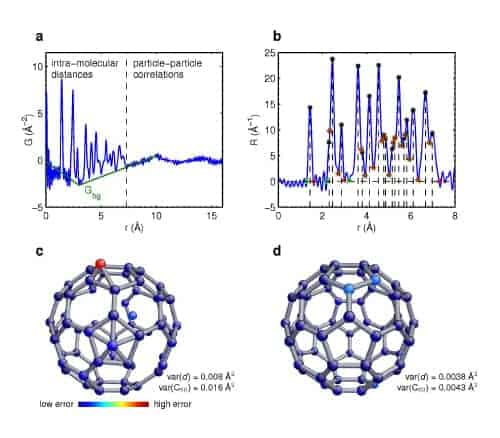
Since nanoparticles do not have long-range crystalline order, it is difficult to use conventional crystallographic methods to look at their structure. But the new algorithm allows researchers to determine the 3D structure of a nanoparticle from 1D X-ray or neutron diffraction data, which is the type of data normally obtained from standard X-ray and neutron diffraction techniques for a powder ensemble of nanoparticles.
There were challenges to overcome along the way though because recreating a 3D nanoparticle – containing around 100 atoms — from a 1D dataset is a computationally difficult problem. This is because traditional algorithms used to build up a 3D picture of a particle, such as the Monte Carlo approach, fail for very small clusters of atoms. The Michigan team therefore decided to try a “cluster build-up” approach instead. This involves proposing “trial structures” and simulating the data for these structures: “how ‘good’ a proposed nanocluster structure is depends on how much its 1D X-ray and neutron diffraction characteristics agree with the simulated data,” says Billinge.
The researchers used their algorithm to solve the structure of a C60 molecule from its “atomic pair distribution function” (PDF), measured using neutron diffraction. PDF data from nanoclusters made up of single elements contains a simple list of the distances between pairs of atoms in the cluster but it does not provide information directly on how those atoms are arranged. The Liga algorithm uses a trial and error approach to recreate the structure using just the distance list as input.
“Our algorithm uses a competition between the clusters like a soccer league,” explains Billinge. “Good clusters get promoted and grow by buying players (atoms); poor clusters are relegated and sell their worst player before competing again at the lower level. The cluster that ultimately wins the top division, the league champion, is the correct cluster. We call it the ‘Liga’ algorithm – well, it sounds better than league.”
Initial applications for the technique will be for inorganic nanomaterials. “It could also be used for studying molecules and clusters in solution,” says Billinge. “By incorporating chemical information and other a priori constraints it may be possible to study quite complex systems, but we don’t yet know how far it will go.”
The team is now extending the Liga algorithm to handle systems containing more than one element, and larger clusters, with a view to applying it to “real systems of physical and chemical interest”.




