Two independent groups of physicists have found a new way to get high-definition glimpses of the innards of living cells. The technique uses X-ray diffraction microscopy on cells frozen below –170 °C, and could pave the way for highly detailed 3D images of cells. This would provide biologists with valuable new information about how the smallest structures in cells interact with one another.
Imaging the tiny structures in cells has been difficult because conventional optical microscopes are limited by the wavelength of light and are unable to pick out fine structures below around 400 nm in size. While electron microscopes can resolve details as small as 3–5 nm, they are unable to penetrate very deeply through biological samples.
The most promising solution is X-ray diffraction microscopy (XDM), which can image whole cells at once and pick out much smaller features, down to around 10 nm. XDM does not use lenses; instead it uses a CCD to measure the intensity and trajectory of X-rays that have been diffracted from the target object. A computer algorithm is then used to reconstruct the image from this diffraction data.
Radiation damage
However, using X-rays on biological samples usually causes significant radiation damage to the cells, which reduces the resolution of any images produced. To get around this problem, previous work has focused on freeze-dried cells, with all water removed from the system. Unfortunately, this process no longer provides a realistic picture of how the cells might appear while alive.
Instead of freeze-drying cells to be imaged, the two teams have plunged cells in their natural wet state into liquid ethane. This freezes the cells very rapidly, which preserves much of their structure while making them more resistant to radiation damage.
“The difference between dehydrated cells and frozen hydrated cells is like the difference between raisins and grapes” Xiaojing Huang, Stony Brook University
“The difference between dehydrated cells and frozen hydrated cells is like the difference between raisins and grapes,” explained Xiaojing Huang of Stony Brook University in New York. “This process does not damage the cells, and keeps them very close to their natural living state, so it gives us a much more accurate picture of what is happening,” he told physicsworld.com.
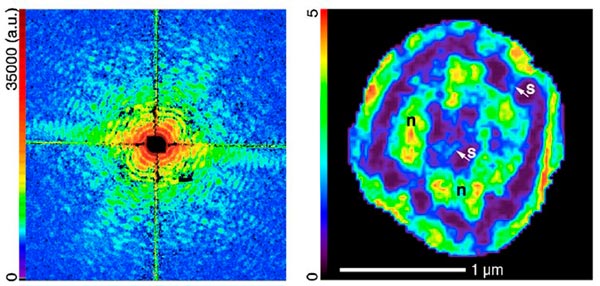
Huang’s group used “soft” 520 eV X-rays to image yeast cells at the Advanced Light Source at Lawrence Berkeley National Laboratory, California, US. At the same time, researchers led by Anders Madsen at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France, used a similar freezing technique and “harder” 8 keV X-rays to image radiation-resistant cells of the bacteria D. radiodurans. In each case, the team was able to attain resolutions of between 25 and 50 nm – enough to discern internal structures of interest, such as possible mitochondria or nucleus regions. Both results are published in Physical Review Letters.
Freezing advantage
The low radiation damage associated with this technique should allow the same cell to be rotated slowly and imaged from many different angles to build up a 3D image.
Huang’s team has already started on this next stage, and is planning to build and implement a system that would automatically collect and produce 3D images in minutes.
“All the work so far indicates that 3D imaging is possible using this process” John Miao, University of California, Los Angeles
“3D imaging is very important because the most interesting cells are relatively thick,” says John Miao, who is also working on imaging cells at the University of California, Los Angeles. “In a 2D-projection you don’t see all the details you would like to, such as internal organelles or possibly larger macromolecules.” “I think this is a very important step in the right direction,” Miao adds, noting that high-quality 3D images may be available sooner than we think. “All the work so far indicates that 3D imaging is possible using this process and the images could even be as high as 10 nm in resolution in the future.”