A multidisciplinary team of physicists and biologists has discovered a new system of proteins that organizes itself spontaneously into a spectacular pattern of whorls and vortices. But unlike similar systems studied in the past, the individual protein molecules move freely throughout the pattern, while the pattern itself remains fixed. The scientists believe that further study of the system could shed light on the collective behaviour of living organisms.
The spontaneous, self-organized collective motion of entities as diverse as flocks of birds and colonies of bacteria have long fascinated humans. Scientists, however, have struggled to understand the underlying mechanisms that allow hundreds, thousands or even millions of individuals to act as one. It is often the case that the dynamics of a group cannot be predicted in a straightforward manner from observing the behaviour of individual members.
Difficult to disentangle
While this collective behaviour in living organisms is fascinating, it is also extremely difficult to model mathematically because of the complicated ways in which living things can interact. These range from one bird hearing another’s distress call to a person being influenced by a friend’s political ideas. It is often difficult to identify all these interactions, let alone to disentangle their influences on a system’s behaviour. Furthermore, conducting interventional experiments can be problematic, as many of the parameters may not be under the experimenter’s control.
Fortunately for scientists, self-organization is not limited to living organisms. By studying often simpler inanimate systems, important insights into collective behaviour can be gleaned. In 2010, two research groups turned their attention to the formation of patterns by protein filaments made from actin, which are driven by motor proteins. These filaments are used by cells to convert chemical energy into kinetic energy by propelling the tiny protein filaments like the outboard motor of a speedboat. Above a certain critical concentration, the researchers discovered that protein filaments on a glass slide organized themselves into various ordered structures such as waves and spirals.
Whorls and vortices
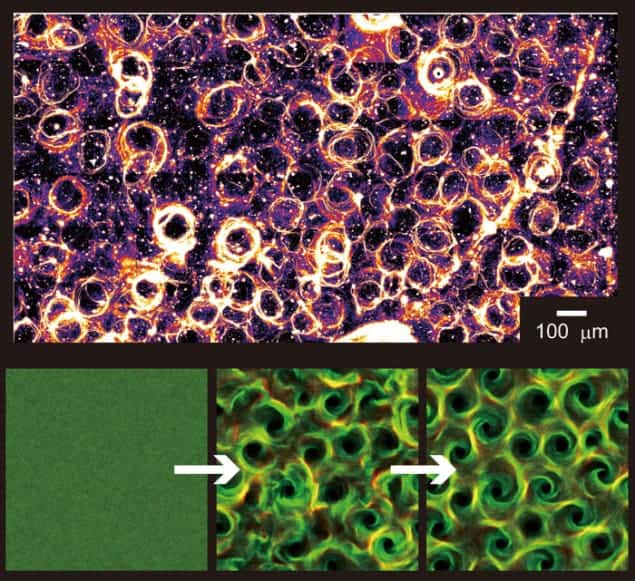
Now, scientists in Japan have looked at a different combination of filament and motor, and found that the proteins then organize themselves into an elaborate pattern of whorls and vortices when placed on a glass plate. The research provides an important new demonstration of complex behaviour arising in a simple system.
The new research is a collaboration involving Yutaka Sumino at the Aichi University of Education, Ken Nagai at the University of Tokyo, Kazuhiro Oiwa at the Advanced ICT Research Centre in Kobe, Hugues Chaté at CEA-Saclay in France and their respective teams. The researchers used a different kind of protein filament called a microtubule and a different type of motor protein called dynein. As with actin, they found that when the density of motor proteins was high enough, the filaments organized themselves into patterns. However, unlike the previous examples of relatively simple patterns, the pattern here was a far more elaborate, repeating “lattice” of adjacent vortices.
Crucially, however, while the established pattern remained stable, a single microtubule was not stuck in one vortex. Individual molecules wandered across the surface of the substrate from vortex to vortex, sometimes rotating clockwise and sometimes anticlockwise, all without disturbing the stability of the overall pattern. Although the researchers were able to construct a computer model of the behaviour, it is not yet clear whether the behaviour is the result of a specific property of the microtubule molecule or whether there may be a more general explanation.
Not frozen-in
Biophysicist Andreas Bausch of the Technical University of Munich in Germany. leader of one of the research groups that originally discovered pattern formation in protein filaments, is fascinated. “These are not frozen-in structures as, for example, we observed last year,” he explains. “You’ve got the constant exchange of material between the vortices and yet they’re stable over very long timescales.”
Tamas Vicsek of Eotvos University in Hungary, an expert on complexity theory and author of an accompanying commentary on the paper describing the work, believes the research marks a significant step forward in the study of complex behaviour in protein filaments. “They just observed a new phase in the study of collective motion that has not been seen before,” he says. “It seemed that searching for new patterns was over, but this shows that the whole system of self-propelled particles is a much richer world than we had assumed.”
Sumino hopes that the work may one day provide a basis for understanding complexity theory more widely. “In our model all you have is a short-range collisional interaction,” he explains. “Depending on the system, sometimes there is more detailed interaction. So if you think about the collective motion of fish, fish have eyesight in one direction only. Such detailed information can be taken into the model later.”
The research is published in Nature.



