
A new type of super-resolution microscopy developed by researchers in Germany, Argentina and Sweden combines the merits of two Nobel-prize-winning techniques, attaining nanometre-scale resolution more quickly and with fewer emitted photons than previously possible.
The resolution limit of traditional optical microscopy is set by the Rayleigh criterion: if two features are separated in space by less than half a wavelength, diffraction will blur the light too much for the features to be distinguished. Super-resolution microscopy techniques surpass this limit by selectively exciting individual fluorescent groups (fluorophores) on molecules while their neighbours remain dark.
Single-molecule microscopy techniques, for which Eric Betzig and William Moerner each shared a third of the 2014 Nobel Prize for Chemistry, switch fluorophores on and off randomly and use the arrival positions of the emitted photons at a camera to reconstruct the location of each fluorophore. However, the photons are diffracted as they escape, leaving some uncertainty about where a single photon originated. Detecting many photons from the same fluorophore can reduce this uncertainty, but to get to single-molecule (nanometre) resolution requires tens or even hundreds of thousands of photons, and most fluorophores degrade (or bleach) before they emit this much light.
Doughnuts of light
Stimulated emission depletion (STED) microscopy, which won Stefan Hell the remaining third of the Nobel prize, uses two beams of light. The first illuminates the sample with a focused beam, turning fluorophores on. The second beam is focused to a doughnut shape in the sample and suppresses the fluorescence everywhere in the focal region – except at the doughnut’s central hole. By scanning the beams jointly over the sample, the spatial distribution of all the fluorophores can be determined. Unfortunately, to suppress fluorescence perfectly except in one single-molecule-sized spot would require a doughnut-shaped beam powerful enough to destroy the sample. Both techniques, therefore, struggle to obtain molecular-scale resolution.
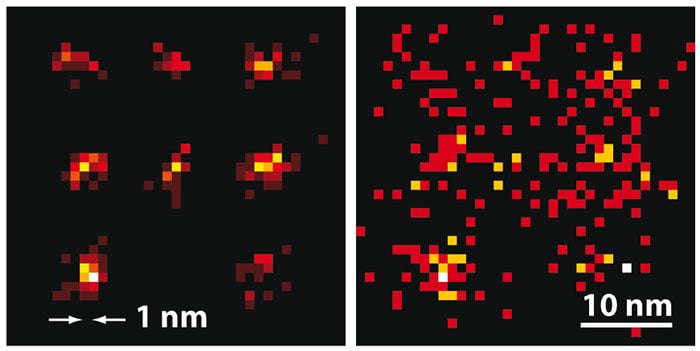
Now, Hell and colleagues at the Max Planck Institute for Biophysical Chemistry in Göttingen and several other institutes have developed a new technique called maximally informative luminescence excitation probing (MINFLUX). They switch individual molecules on as in single-molecule microscopy, but for determining the molecules’ position, they scan a single, doughnut-shaped beam across the sample, as in STED microscopy.
When their photodetector records a signal, the researchers know that a fluorophore is nearby. As the beam is doughnut-shaped, with zero intensity at its centre and increasing intensity further away, the intensity of the fluorescence can reveal the intensity of the light incident on the fluorophore and, therefore, how far the fluorophore is from the focus of the beam.
Beam structure
By aiming the beam at four points near the fluorophore and recording the fluorescence intensity each time, the researchers can easily work out how far the fluorophore is from each point and deduce its location with nanometre-scale precision. “In single-molecule microscopy, the co-ordinate system is given by the pixels of the camera,” explains Hell, “Here, it’s the structure of the beam that defines position.”
It’s a very clever idea and it’s remarkably simple
Adam Cohen, Harvard University
The researchers used MINFLUX to map the positions of fluorophores tagged onto specially shaped sequences of DNA called DNA origamis, and thus to calculate their shapes. In the first experiment, using fluorophores separated by 11 nm, the researchers needed only 50 s of imaging time to locate all those that emitted at least 500 photons in total with an average spatial uncertainty of 2.1 nm. Then, using fluorophores only 6 nm apart, they identified those emitting more than 1000 photons to within around 1.2 nm in 120 s.
Twice as good
These precisions are both twice as good as the maximum precisions theoretically achievable using single-molecule microscopy with the same numbers of photons. The researchers also tracked single molecules inside living bacteria – something only possible because of the faster speed of MINFLUX.
“MINFLUX is a truly remarkable breakthrough with a highly innovative method design,” says Xiaowei Zhuang of Harvard University in the US – one of the inventors of single-molecule super-resolution microscopy. “It’s amazing ability to localize molecules at an ultra-high precision with such a low photon number will greatly benefit our understanding of molecular interactions inside cells.”
Adam Cohen, also at Harvard, agrees: “It’s a very clever idea and it’s remarkably simple,” he says. “People have been trying to track molecules for a variety of different applications for the last 15 or so years, and this relatively simple concept will improve that quite a bit, at least under some circumstances.” He sees a potential issue with the use of the technique in more complex cellular structures, however: “If you start to get stray photons coming not from your molecule but from other sources, those stray photons will throw off the precision of the tracking,” he explains. “As you get to having more and more fluorophores in your sample, it becomes increasingly difficult to ensure that all but one of them will be off.”
The research is described in Science.



