By applying deep-learning techniques to a set of phase-contrast microscopy images, Japanese researchers have been able to identify the nature and origin of different cancer cells with 96% accuracy. This approach could lead to better cancer treatments (Cancer Res. 10.1158/0008-5472.CAN-18-0653).
The researchers, from Osaka University, used a convolutional neural network (CNN), a common scheme used in deep learning, to analyse the images. CNNs work by applying to the input image a set of connected filters and mathematical functions that, similarly to neurons, can be trained to extract specific features. In medical imaging, CNNs are modelled on the human visual system, with low layers that capture fine details such as edges, and higher levels that capture complex features reflecting the whole image.
Training AI to spot tumours
The idea of applying artificial intelligence (AI) to medical images for clinical purposes is not new. But many have questioned the ability of AI to distinguish the fine details that differentiate tumours that have developed resistance to common therapies.
Identifying the type of tumour and its origin is indeed a key step towards personalizing treatment for a specific patient. There is, for example, no need to expose a patient to radiotherapy if their tumours are radioresistant — as this could be both detrimental to them and a waste of time. Until now, however, tumour classification was mainly performed by visual inspection, which made the process time-consuming and prone to human-error.
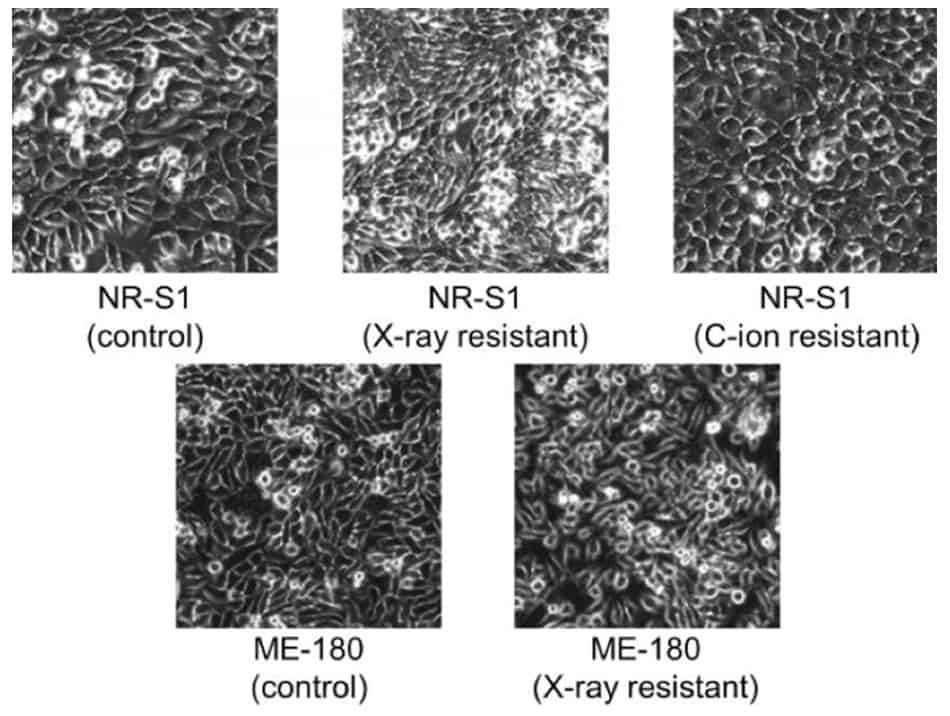
In this study, the researchers designed the CNN to classify cells into five categories: untreated (control); X-ray-resistant or carbon-ion beam-resistant mouse tumours (NR-S1 type); untreated and X-ray-resistant human cervical tumours (ME-180 type). They subsequently trained the CNN with a database of 8000 phase-contrast microscopic images containing these types of cells and validated it using an additional 2000 images.
In need of a comprehensive database
The network was able to correctly identify 96% of the cells in the validation dataset, although it had more difficulty recognizing the human cell lines. This was especially the case for X-ray resistant cells, which had only a 91% rate of success, compared with 99% for the other types of tumour tested. This pattern was confirmed when all the 4096 features extracted by the CNN for each image of the two datasets were collated to form a 2D map: while the three clusters of cells originating from mice were distinct from each other, the two clusters of human cells were relatively close to each other.
These results are a stepping stone towards a more ambitious design. In the future, the team hopes to train the system on more cancer cell types, with the eventual objective of establishing a universal system that can automatically identify and distinguish such cells.