Detecting a brain tumour at the earliest possible stage enables faster treatment and safer surgery, which are essential to improve the patient’s chance of a good clinical outcome. But brain tumour diagnosis is a difficult task, as common symptoms such as headaches or memory change are not specific to cancer. As such, many tumours remain undetected until they are larger or of a higher grade. A research team in the UK has now demonstrated that spectroscopic liquid biopsy can detect both small and low-grade gliomas – and could increase the likelihood of early diagnosis.
Liquid biopsies are a minimally invasive diagnostic tool in which small samples of blood are analysed. For cancer diagnostics, most liquid biopsies detect genetic material such as circulating DNA. But early-stage tumours can have extremely low levels of cancerous genetic material in the blood, and for brain tumours, the blood–brain barrier creates further limitations.
The researchers are developing an alternative approach. Instead of detecting specific genetic material, they analyse the molecular composition of a blood sample using attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy combined with machine learning algorithms.
“The spectroscopy is used as a broad assessment of the entire composition of the blood sample, which contains over 20,000 molecules,” explains first author Ashton Theakstone from the University of Strathclyde.
Serum sample studies
In their latest study, described in Cancers, Theakstone and colleagues examined 177 blood serum samples from patients with varying sizes of brain tumours, including 90 patients with high- and low-grade gliomas, as well as 87 asymptomatic controls. They measured the tumour volumes using MR imaging and divided the patients were into two groups depending on their MRI parameters (T1-weighted with contrast enhancement, or T2-weighted/FLAIR).
The researchers performed the liquid biopsies by depositing 3 µl of each patient’s serum onto an optical sample slide and collecting nine spectra per patient, which typically took 15 minutes. They collected spectra in the wavenumber range 4000–450 cm-1, with spectral analysis focused on the fingerprint region (1800–1000 cm−1).
The team first used principal component analysis (PCA) to examine spectra from the T1 group, all of whom had grade IV glioblastoma tumours, and from control patients. PCA provides clear visualization of any variation between these datasets and identifies important wavenumber regions within the spectral data. The analysis revealed a distinction between tumours and controls, which the researchers used to determine the wavenumber bands responsible for this separation.
“Any variances within particular wavenumber regions correspond to certain functional groups that are known within the literature,” Theakstone explains. “For example, the region between roughly 1700 to 1500 cm-1 corresponds to the amide I and amide II of proteins, therefore variances within this region relate to fluctuations in protein content within the blood.”
Cancer classification
Next, the team examined the ability of three classification models (random forest, partial least squares-discriminant analysis (PLS-DA) and support vector machine) to differentiate these high-grade tumours from controls. For each model, the data were split into a training set, used to identify biosignatures, and a test set.
The PLS-DA model had the greatest predictive ability, with a sensitivity of 98.5%, specificity of 95.1% and balanced accuracy of 96.8%. The team repeated this process with the T2/FLAIR group, which included mostly low-grade (grade II) tumours and some grade III tumours. Again, PLS-DA performed the best in differentiating tumours from controls, with a sensitivity of 88.7%, specificity of 94.7% and balanced accuracy of 91.7%.
To minimize sensitivity and specificity errors whilst also minimizing analysis time, the researchers repeated each classification 51 times. They note that the first iteration for each classification model gave correct predictions for all patients within the T1 cohort and the majority of the T2/FLAIR group.
Significantly, the model correctly identified a patient with a tumour size of just 0.2 cm3 as a cancer patient for each iteration and for all nine of their recorded spectra. “Therefore, we can confidently state that this model will identify tumours as small as 0.2 cm3 with 100% success rate in this particular example,” says Theakstone.
Focused ultrasound tackles brain tumours in a myriad ways
The researchers conclude that their spectroscopic liquid biopsy shows great promise as a potential diagnostic tool for early diagnosis of brain tumours. Importantly, and unlike other liquid biopsies, this approach appears insensitive to tumour volume. They note that the test could be used to fast track patients who need medical imaging, and are continuing this research to deliver an early-stage triage tool for brain cancer detection.
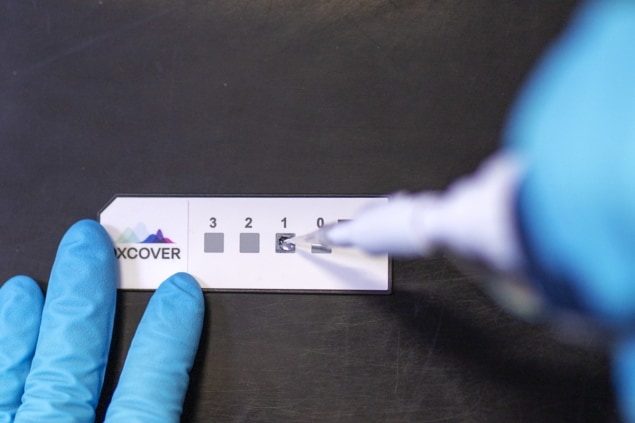
Furthermore, Glasgow-based Dxcover, a spin-out from the University of Strathclyde, is commercializing an early detection platform based on the spectroscopic liquid biopsy technique.
“This breakthrough is a watershed moment in the development of early cancer detection,” says Matthew Baker, Dxcover’s chief technical officer and co-founder, in a press statement. “The study demonstrates the effectiveness of our Dxcover Brain Cancer Liquid Biopsy at detecting even the smallest brain tumours, which is great news for the care of future brain cancer patients, increasing treatment options and potentially extending life expectancy.”