A computational model developed by scientists and engineers in the US describes different ways in which gene products are capable of performing specific biological functions. Their results also help explain how certain ways of performing specific functions may have enabled complex multicellular organisms to evolve more readily (PNAS 10.1073/pnas.1815912115).
Upon receipt of particular signals as inputs, biological systems respond by performing particular functions. Two main mechanisms mediate functional specificity in biology: “lock-key” interactions, as for example when an enzyme is the lock and its substrate is the key; and weak cooperative interactions (WCI). In the first class, the substrate can fit into the lock (the enzyme’s active site) only if it is perfectly right in term of size and other properties.
Increasing evidence suggests that the second class, WCI, is more prevalent in higher organisms. For example, exciting recent insights suggest a quarter of human genes encode proteins that can functionally interact through WCI in executing critical tasks. In the context of immunology, WCIs are important for how the immune system can differentiate different antigens (specificity). Functional specificity mediated by WCI can lead to some cross-reactivity, such as when the same immune cell can “recognize” different antigens. However, the factors that drove the evolution and selection of WCI for mediating functional specificity in multicellular organisms is yet to be understood.
Therefore, researchers are motivated to learn more about how WCI underlies functional specificity, not only because of its far-reaching implications for biology but also due to its potential contributions to dangerous pathologies when cross-reactivity is misregulated.
In silico simulation of evolution
To gain insights into the driving forces that mediate biological specificity in complex organisms, researchers from Massachusetts Institute of Technology and the Ragon Institute of MGH, MIT and Harvard, led by Arup Chakraborty and Phillip Sharp, developed a model where a population of organisms evolves as the number of tasks that must be carried out with functional specificity increases. In other words, their model is based on interactions between the organisms’ gene products and the tasks that they must perform. Indeed, their model is inspired by the interactions that lead to protein–protein recognition.
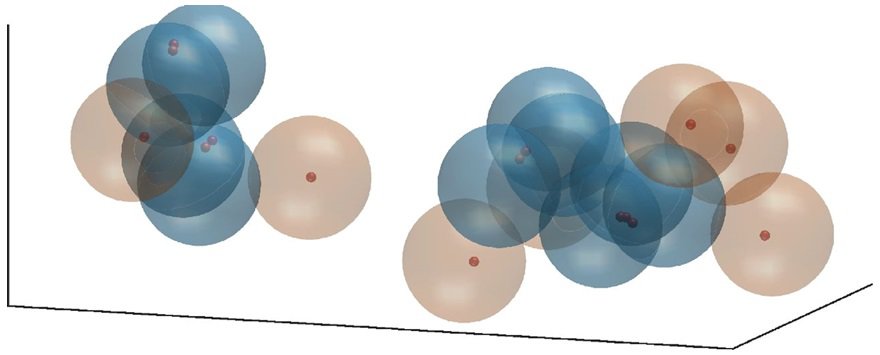
Briefly, the model represents a space composed of gene products of organisms and the tasks that need to be performed with specificity for the organisms to function properly. Gene products and their tasks are defined as positions in a characteristic space, with axes that define different properties of the tasks and the gene products. Well-matched gene products and tasks are closer to each other in characteristic space, and if they are close enough, the task is considered to be performed specifically by the corresponding gene product. Similarly, well matched gene products that can perform a task by their cooperative action are close to each other in characteristic space.
Mapping evolutionary dynamics
The researchers found that when there are very few tasks that must be performed specifically for organisms to function properly, the lock-key mechanism is dominant. As the number of tasks that had to be performed with functional specificity increased, WCI naturally evolved as a mechanism for mediating functional specificity and became increasingly prevalent. This result led the researchers to wonder whether there is a link between the emergence of WCI and becoming more evolvable – i.e., the evolvability of multicellular organisms.
Mutations, gene loss and gene duplication underlie evolution. It is thought that more evolvable systems require fewer number of mutations to create new phenotypes in a shorter time period. Therefore, to explore the relationship between WCI and evolvability, the researchers compared simulations where emergence of WCI was not allowed to those where WCI was allowed.
They found that the model allowing the evolution of WCI exhibited shorter response times and required fewer mutations to respond to new tasks. This enables higher-order organisms to efficiently perform new tasks in addition to maintenance of old tasks. Thus, the authors suggest that WCI has been repeatedly positively selected in increasingly more complex multicellular organisms.
These findings may have implications beyond protein–protein or specific enzyme–substrate interactions and immunology. One example is the nervous system. “It may also be a characteristic of how computational machine learning algorithms trained on large datasets to predict specific outcomes could be adapted to predict new outcomes,” the authors speculate.