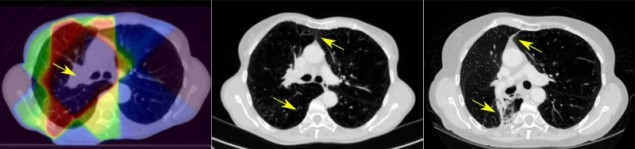
Radiation therapy of lung cancer can cause toxic side-effects to healthy lung tissue, known as radiation-induced lung damage (RILD). RILD can significantly impact patient quality-of-life post-treatment, but with historically poor prognoses of lung cancer, long-term RILD reporting is piecemeal, with only a few subjective and unreliable scoring systems available. Recently, however, improvements in lung cancer survival have triggered interest in more rigorous RILD assessment.
To establish a standardized methodology for RILD scoring, medical physicists and engineers from University College London (UCL) teamed up with clinical oncologists and thoracic radiologists from Guy’s and St. Thomas’s NHS Trust and Royal Brompton Hospital.
The multidisciplinary team analysed CT scans of cancer patients before and after treatment, and developed software to semi-automate quantification of 12 identified biomarkers of RILD. This is the first time that RILD has been quantified using a broad spectrum of radiological findings (Int. J. Radiat. Oncol. Biol. Phys. 10 1016) .
“As we start to get long-term survival, looking at the lasting damage to the patients becomes more important,” says first author Catarina Veiga, research associate at UCL. “We wanted to develop methods to quantify this in a very objective manner.”
Spot the difference
In the first stage of the study, clinicians carefully examined CT scans of 27 non-small cell lung cancer patients who had taken part in a phase I/II clinical trial of isotoxic chemoradiation. By aligning the baseline images with those taken 12 months post-treatment, they identified common changes.
“All patients at 12 months had some type of lung damage, but the degree of change could be different,” explains Veiga. “Some just have a bit of lung volume loss, while others have extensive parenchymal damage, pleural effusion and anatomical distortions.”
Previous to this study, the main RILD marker recognised was parenchymal changes – scarring or inflammation that can be easily seen on CT imaging as “bright” regions within the lung. Here, the clinicians identified three main categories of RILD – parenchymal, pleural and anatomical.
In the pleura – the area between lungs and chest – inflammatory changes were observed. And the clinicians noted that as treatment scars shrunk, they distorted the lung’s anatomy, so reducing lung volume. From these observations, the team settled on 12 measurable biomarkers of RILD.
Automating for objectivity
At this point, the engineers stepped in to quantify the biomarkers and establish an image analysis pipeline in a modular and semi-automated fashion within MATLAB. The pipeline was validated by comparing results to the original visual observations.
Of the 12 biomarkers, 10 showed a significant change, and statistical analysis highlighted that biomarkers were independent of one another. This made the authors confident that they have a system that can accurately report RILD, and they plan to make it fully automated and freely available to the community in the future.
“By automating all of this, we can speedily process the results from trials and clinical practice, and have evidence of how the different treatments we are using may or may not impact the long-term outcome,” says Veiga.
Technical challenges
The scientists note that there are some technical limitations to their methodology. Imaging protocols are of key importance, with inconsistency easily introduced between scanning time points. For example, variations in a patient’s breathing during imaging could influence biomarker measurement.
“For this particular clinical trial, the majority of patients had consistent imaging, but in routine clinical practice it is likely that this will become more of a challenge,” says Veiga.
Another technical consideration that the team is trying to address is the use of manual segmentation methods. Although most stages of the pipeline are automated, images were sometimes manually segmented to calculate the biomarkers. This is a laborious process that could introduce subjectivity to the results, and so Veiga and colleagues are currently running a pilot study that fully automates segmentation.
Improving life quality for survivors
“Once we are able to fully automate the system then we want to start looking at how the damage evolves over time,” notes Veiga, who is planning to examine the three, six and 24 month images that they have from the same clinical trial. “We are also interested in looking at the relationship between the identified damage, radiation dose delivered and clinical outcomes.”
Ultimately, Veiga hopes that the multidisciplinary team has created a tool that can improve clinical practice and be used in future trials to find the critical radiation dosage that clears tumour cells with minimum toxicity.