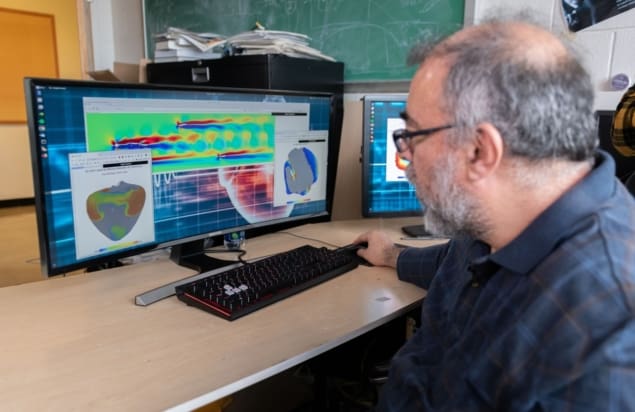
Modelling the complex electrical waves that cause heart arrhythmias could be key to understanding and treating these abnormal heart rhythms. Until now, however, real-time modelling of cardiac dynamics within millions of interacting heart cells required access to powerful computer clusters and supercomputers, putting it out of reach for most physicians.
To make cardiac modelling more accessible, a team of US researchers has used graphics processing chips and software that runs on standard web browsers to move high-performance cardiac dynamics simulations onto less costly computers, and even high-end smartphones. This advance could enable clinicians to use 3D modelling data to design specific therapies or prevention strategies for their patients, and help scientists study a particular drug’s effect on heart arrythmias (Science Advances 10.1126/sciadv.aav6019).
“Being able to do real-time simulations in three dimensions could open the door to clinical applications where we could actually obtain patient geometries and solve these equations in the cells that are packed into the heart,” says Elizabeth Cherry from Rochester Institute of Technology. “We could see applications in the clinic that could individualize treatments on the basis of their specific heart geometries. We could actually test possible therapies to see what would work for each patient.”
Key to this development is the use of graphics processing units (GPUs) designed for gaming applications, which were developed to help computers display graphics and video. High-end smartphones can have up to 900 GPU cores, while high-end graphics cards for laptop or desktop computers may contain more than 5000.
“Over the past several years, GPUs have become really powerful,” explains Flavio Fenton from Georgia Institute of Technology. “Each one has multiple processors, so you can run problems in parallel like a supercomputer does. As many as 40 or 50 differential equations must be calculated for each [heart] cell, and we need to understand how millions of cells interact.”
To allow the simulations to run on any GPU, Georgia Tech’s Abouzar Kaboudian developed a versatile programming library that enabled the team to develop programs in WebGL that can run through common web browsers.
“If you have access to the Internet and a modern web browser like Firefox or Chrome, you can just go to a web link and the simulation will start running on the graphics card of your computer,” says Kaboudian. “Any problem that can be parallelized can run on the library that we have created. It will accelerate simulations on any computer by several hundred times.”
The researchers have developed ten different models based on their WebGL programming, and are planning to make the tools available for other researchers to use. Future enhancements will include the ability to run simulations on more than one GPU card to achieve even higher computational speeds.
“Models that might have been accessible to only a handful of researchers in the world will now be available to many more groups,” says Fenton.



