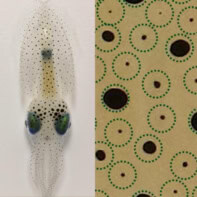
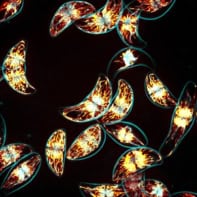
This image may look like a collection of novelty spaghetti shapes but these detailed figures are in actual fact made from assemblies of DNA strands as imaged by an atomic force microscope (AFM). It is the demonstration of a pioneering technique, developed at Harvard University in the US, for engineering complex nanoscale structures from a set of DNA “tiles”.
The concept of fashioning nanoscale structures out of individual strands of DNA has been developed over the past three decades. The field has gathered momentum since the emergence in 2006 of a technique known as “DNA origami”, which enables scientists to fold long single strands of DNA into a wide range of predetermined shapes. These resulting nanostructures can be used as scaffolding or as miniature circuit boards for precisely assembling components such as carbon nanotubes and nanowires.
But despite early applications and the great promise of the origami technique, there are limitations with the production process relating to time and expense. In coercing DNA into folding, scientists must attach several hundred staples to the area surrounding the single strands. Each shape requires a different set of staples, which cost approximately £640 each, and the entire process from design through to fabrication typically takes a week.
Molecular canvas
In this latest research, Peng Yin and colleagues at Harvard University have developed an alternative technique for DNA engineering, based on an earlier manipulation approach that dates back to the 1990s. Instead of starting with one long strand of DNA and folding it, Yin’s team takes a number of shorter single strands of DNA and arrange them into a “molecular canvas” by controlling their local interactions.
It works because the DNA has been programmed to have specific sequences of bases: each single strand comprises 42 bases divided into four sequences of 10 or 11 bases. Single strands are then folded up into rectangles, or “tiles”, that are mixed together with other tiles in an environment where each sequence will only be attracted to one other complementary sequence. In this way, researchers can create specific shapes through the selection of sequences.
Reporting its findings in Nature, Yin’s group demonstrates its technique by presenting these AFM images of 100 distinct shapes including the capital letters of the Latin alphabet, emoticons and astrological symbols, with each shape taking just one hour to produce. These images have been enlarged and their real sizes are 150 nm × 150 nm. Each image was created by selecting a subset of DNA strands from a set of 310, where a further 1396 strands were required for seal the edges of the shapes to prevent them from aggregating. This set of tiles cost roughly £4500, but the researchers estimate that it could make 2 × 1093 possible shapes.
Drug delivery
“Strands are programmed to have sequences that encode local interactions – like every one of them remembers which one is to its left, right, top and bottom – with this, strands can easily find their position within the big molecular canvas, collectively,” Yin told physicsworld.com. Yin thinks that the technique could be applied to a large number of possible applications, including the creation of nanoscale drug-delivery vehicles. In the longer term, Yin believes that the technique could be used to build scaffolds and machinery inside the body that would interact with proteins and other cellular components in a more programmable fashion.
Ebbe Sloth Andersen, a biophysicist at Aarhus University in Denmark, agrees that the ability to create multiple shapes from the same set of DNA provides a distinct advantage for this technique over DNA origami. “The technique will, in general, accelerate the research field of DNA nanotechnology by making the design of DNA nanostructures cheaper, faster and easier than before,” he says. Anderson has concerns, however, about the reliability of the technique in its current state. “The main limitation is the yield of the new technique in the 6–40% range, which does not compare with that of DNA origami where 95% yield is regularly obtained.”