A team at Massachusetts Institute of Technology and Arizona State University has developed a program that can convert free drawn shapes to 2D DNA nanostructures, also known as DNA origami. This algorithm can autonomously determine the required sequences, simplifying the process of designing these highly detailed assemblies.
DNA origami is a technique that creates nano-sized 2D and 3D structures made of DNA. These shapes can be particularly useful in nanotechnology for producing complex structures. The approach uses the ability of DNA bases to pair with each other to form a large “scaffold” sequence of DNA and hold these together with smaller “staple” sequences.
Until recently, designing DNA origami required researchers to manually work out the sequences required, as there were limited options for automating this long process. However, Mark Bathe and his team have produced an algorithm that is capable of calculating the strands needed to assemble a desired shape. This removes the need for expert knowledge when designing DNA origami (Science Advances 10.1126/sciadv.aav0655).
Exploring DNA designs
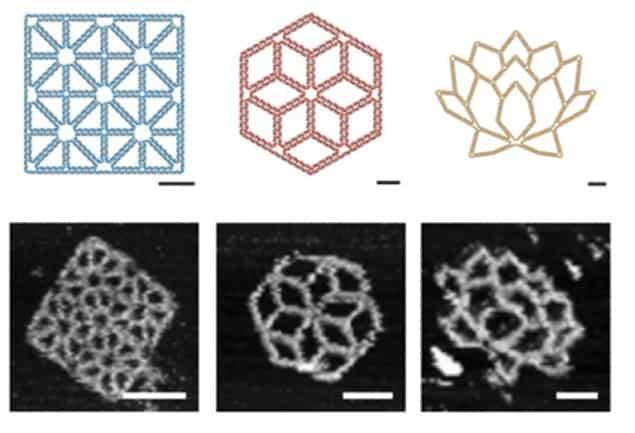
To test the abilities of the program, the researchers designed several structures for it to recreate. They used the sequences that the program generated to create the origami and examined the resulting shapes using atomic force microscopy (AFM). The algorithm was capable of producing many different shapes with varying edge lengths, angles and internal meshes. They were even able to produce highly complex and interesting DNA origami, including a lotus flower shape.
The program works by determining the minimum number of staple sequences needed to adhere to the scaffold and form the desired shape. It searches every possible place where the staples can cross over the strands and creates pathways between these possible crosses, allowing researchers to compute the optimal structure. The sequences can then be simply assembled by placing all the strands in solution and using heat to form the structures.
One of the advantages of this software is that edges within the structures do not have to be a specific length, which unlocks a wide variety of possible shapes. Previously, these lengths needed to be a whole number of double helical turns to ensure the origami was flat. This design flexibility is achieved by taking advantage of a new technique that uses DNA bases that do not have a pair on the scaffold strand to bridge the gap.
Opening the field
The program, called PERDIX, is available to researchers as an online tool and standalone software. The team hopes that this technique will allow a greater variety of researchers to develop DNA origami.
“The fact that we can design and fabricate these in a very simple way helps to solve a major bottleneck in our field,” says Bathe. “Now the field can transition toward much broader groups of people in industry and academia being able to functionalize DNA structures and deploy them for diverse applications.”