In animals, plants, fungi or any other eukaryotes (organisms whose cells have nuclei), transcription of DNA into RNA cannot function without a protein called DSIF, but little is known about the exact process enabling it. Carrie Bernecky, Jürgen Plitzko and Patrick Cramer from the Max Planck Institute for Biophysical Chemistry and Max Planck Institute of Biochemistry managed to image DSIF in action while the protein was interacting with RNA Polymerase II (Pol II). These images reveal the structure and function of DSIF, a modular protein that performs a whole range of tasks including holding and positioning DNA and RNA, regulating timing of transcription and subsequent RNA processing for translation.
In the study, the authors show that DSIF consists of two loops, which wrap around the DNA and RNA strand respectively, and a stabilizing domain (Nature Structural and Molecular Biologydoi: 10.1038/nsmb.3465). The protein keeps all molecules involved in transcribing genes into RNA in place: it positions incoming DNA and outgoing RNA, and recruits factors for RNA processing. By physically contacting all key players, including the polymerase itself, it exerts its influence. The positioning of nucleic acids is done by forming clamps, like rings, around the DNA and RNA.
Beyond the resolution of light
To capture images of this process, Bernecky and her colleagues resorted to cryo-electron microscopy and X-ray crystallography. Instead of light, these techniques use electrons and X-rays, respectively, allowing for incredibly high resolution. The much smaller wavelengths of electrons and X-rays overcome the resolution limit that can be achieved with light.
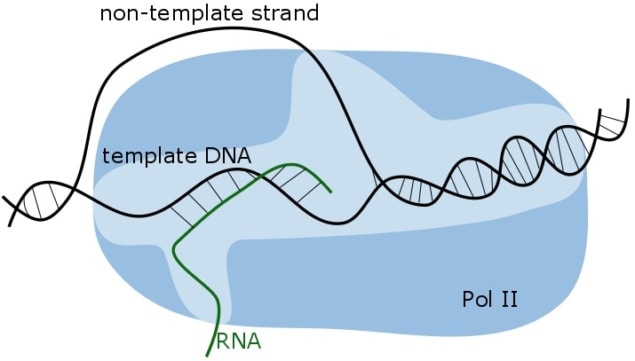
The experiment highlighted DSIF’s interaction with the outgoing RNA that keeps this RNA in the exit channel of Pol II. If this was not the case, the produced RNA would compete with the non-template DNA strand for the template DNA strand and destabilize the interaction between RNA, DNA and polymerase. As a result, transcription would stop.
DSIF pauses and un-pauses Pol II
Another noteworthy feature is the involvement of DSIF in pausing Pol II and releasing it from pause positions. The pausing regulates the rate of transcription and ensures that genes are expressed synchronously, for example to form new tissues during the development of embryos. This can be achieved by un-pausing Pol II simultaneously in many neighbouring cells. Whether DSIF initiates pausing or un-pausing depends on its phosphorylation state.
The DNA clamp, as observed by Bernecky and her co-workers, is important for maintaining stability of the elongation complex. Indeed, without it, Pol II dissociates from the template and transcription pauses. The RNA clamp of DSIF contributes more actively to the regulation of pausing, mediated by phosphorylation. This makes this RNA clamp a major developmental regulator.