Electron tomography is a powerful way to image materials with extremely high resolution but the method cannot provide 3D images on the atomic scale for several reasons. Now, researchers in the US are saying that they may have overcome some of the technique’s limitations by taking a new approach to the problem. The advance could be a boon for those imaging nanomaterials, including biological samples.
Being able to visualize how atoms are arranged in materials has played a crucial role in the evolution of modern science and technology. Crystallography techniques have long been employed to reveal 3D atomic structures and scanning probe microscopes can determine surface structures on the atomic scale. Electron microscopes, for their part, can routinely resolve atoms in 2D projections of 3D crystalline samples.
However, currently there is no direct way to determine the 3D structure of samples on the atomic scale without first knowing the material’s lattice structure and assuming that the atoms fit rigidly on that lattice. These conditions do not apply to a nanoparticle, which can have a different lattice structure than a bulk sample of the same material.
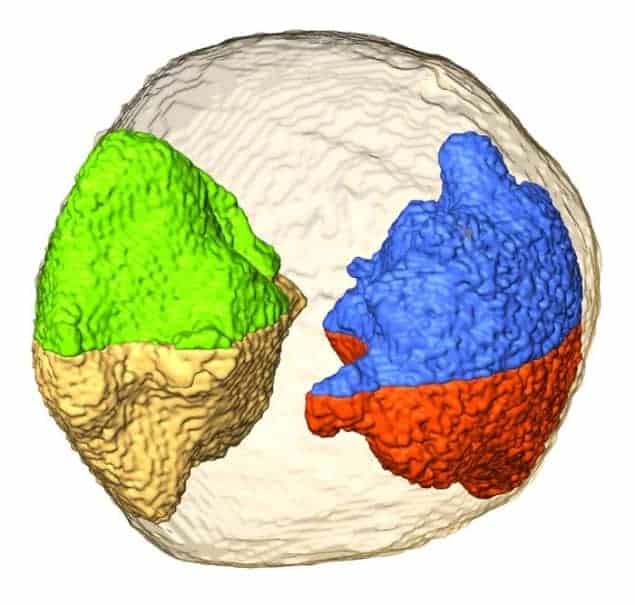
Now, Jianwei Miao and colleagues at the University of California, Los Angeles (UCLA) and the Lawrence Berkeley National Laboratory say that they have performed the first experiments in which they can directly image 3D local structures without relying on a priori structural information. “We have succeeded in observing individual atoms inside some regions of a gold nanoparticle 10 nm in size and identifying several grains therein at a resolution of just 2.4 Å in three dimensions,” Miao says. “This has never been done before.”
Overcoming old challenges
One major drawback of conventional tomography is that samples have to be tilted a number of times and images taken at each tilt to build up the overall structure. But the problem is that prolonged exposure to the electron beams used in the microscope damages the sample and therefore limits the number of projections possible from a single object. Another disadvantage of the technique is that it is technically challenging to align the sample along a common axis and then tilting it with respect to this axis with atomic-level precision. Finally, samples cannot normally be tilted beyond ±79°, which means that data cannot be acquired from the “missing wedge”.
Miao and colleagues have now overcome all of these problems by combining a new alignment method with an iterative tomographic reconstruction technique that uses an annular dark-field scanning transmission electron microscope (ADF-STEM).
The team says that it has been working towards this goal since 2005, when it decided to improve on conventional tomography by developing an “equally sloped tomography” (EST) technique. This method allows the researchers to tilt their samples by small, and as the technique’s name suggests, equally sloped, increments and perform 3D reconstructions on them using a Fourier-based iterative algorithm. The EST helped to significantly reduce the required number of projections from a given sample, which automatically meant less beam damage, while maintaining the same image resolution, image contrast and image quality.
“In our new work, we have combined this EST method with another new projection alignment approach, which relies on the centre of mass of a particle, to image a 10 nm gold nanoparticle in 3D at the atomic scale using only 69 projections,” explains Miao.
According to the UCLA researchers, their general method could, in principle, be used to determine the 3D local structure of crystalline, polycrystalline and even disordered nanomaterials at the atomic scale. It might also help to improve the resolution and image quality when performing electron tomography of biological samples.
The work is presented in Nature.