Super resolution localization images typically require hundreds or thousands of frames to provide a single reconstructed coordinate map of molecular positions. This takes time to acquire, and therefore limits temporal resolution – how fast you can image. Here researchers from Paris use deep learning – a kind of machine learning/artificial intelligence that uses neural networks to massively accelerate the process.
How it works
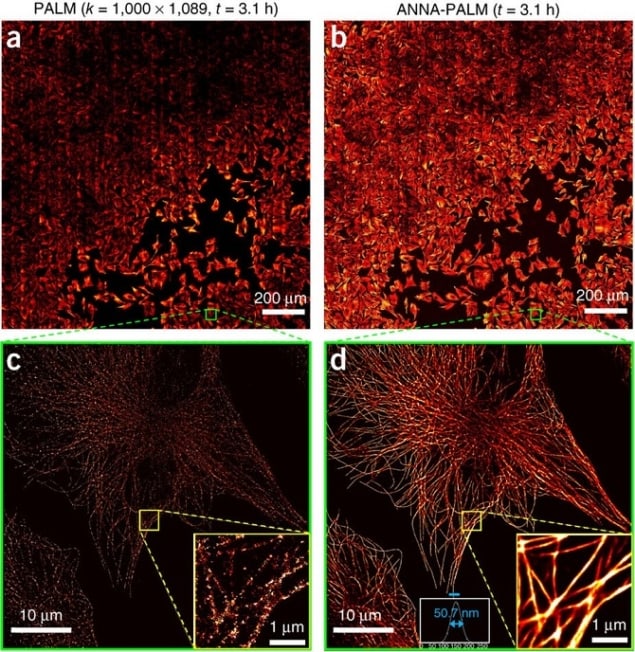
ANNA-PALM (artificial neural network accelerated-photo activated localization microscopy) is based on the idea that a computer can predict the structure of a biological entity from sparse data if it has enough prior information about its expected structure. Training the system requires two bits of data. First, the system needs high-density training data of the structure, to represent its true nature. This can be conventionally acquired using Photoactivated localization microscopy (PALM) or other types of microscopy such as Stochastic Optical Reconstruction Microscopy (STORM) or DNA points accumulation for imaging in nanoscale topography (DNA PAINT) – which all use thousands of frames to get extremely dense structural maps. The second requirement is very low-density input data from the same cell, mirroring the number of frames you want to use for the real experiment. The fewer the frames, the higher the temporal resolution.
The artificial neural network (ANN) is used to recover approximations of the dense training data from the under-sampled data, and its success is measured using a ‘loss’ criterion that takes into account various components of the structure. Once this criterion reports reliable reconstruction of the dense image from the under-sampled image, the ANN is trained. By using a widefield image of each cell of interest in conjunction with the reconstructed ANNA-PALM image, the authors built in an error detection method inspired by a technique called SQUIRREL but using neural networks, to identify how well the software has reconstructed the structure.
After training the ANN on multiple datasets, it can be used to predict the structure of dense images based only on under-sampled images from experimental imaging data. These dense reconstructions, the product of the trained neural network, are therefore new data. They approximate the real situation had the sampling met the Nyquist requirement, where for a given resolution unit there must be at least two independent localizations. (In practice this could refer to the hypothetical ability to label every single G actin protein within an actin fibre, for example.)

Computers ‘learn’ how nanoparticles scatter light
What it can do
The researchers report that the technique is even able to pick up changes in the network due to drug perturbation – a major question if the goal of an experiment is to compare structural networks in multiple experimental conditions. It may therefore be possible to use sparse data to reconstruct more complex structures, such as actin networks that differ in separate parts of the cell – for example the dense actin in the leading edge compared with the stress fibre like actin in the lamella of T cells or dictyostelium. However, careful experimental planning would have to be included in situations where the status of the structure in condition B is completely unknown. Using a few conventionally acquired super-resolution images obtained from a high number of frames is one way of validating this technique.
Clear advantages of the technique exist. The first is its application to high throughput super resolution microscopy. The high temporal resolution afforded by ANNA-PALM means that you can practically image many more cells in a single day. By using an automated imaging system, Ouyang et al. were able to obtain super resolution images of microtubules in more than 1000 fixed cells in a single day. Each cell only required 10 seconds of imaging time to produce a reliable map of microtubules. Transforming a sparse localization image into a dense super-resolution image using ANNA-PALM takes less than 1 second per field of view.
Applying ANNA-PALM to live cell microscopy in conjunction with automated imaging systems, could reduce human bias during cell picking, decrease phototoxicity and increase the technique’s applicability to fast moving cells such as leukocytes.
There are other approaches that also aim to achieve higher temporal resolution. For live cell super-resolution microscopy, some researchers in the field are trying to increase temporal resolution by engineering bright fluorescent proteins that provide sufficient signal for 1 millisecond frame rates. Other researchers are attempting to increase temporal resolution by decreasing the number of required molecules per reconstructed frame to extract meaningful statistical data about protein clusters. Bayesian statistics allows researchers to successfully quantify very sparse datasets, but focuses on the biological phenomenon of protein clustering. Structures, as opposed to clusters, are notoriously hard to capture and quantify by super-resolution localization microscopy, since extracting meaningful quantitative data requires the full structure.
This technique gives researchers access to maps of entire fibre networks (actin, microtubules etc) and other structural components within cells (microtubules, nuclear pores, spectrin repeats etc) with only a few frames of acquisition data. When combined with multiplexed imaging, quantitative analysis techniques, and advances in fluorophore engineering as mentioned above, ANNA-PALM will be a useful technique to elucidate the role of such structures, and how they interact with signalling networks inside living cells.
Full details of the work can be found at Nature Biotechnology.