Can we map all the information being circulated in the human body, and would doing so be any use? Jon Cartwright explores the emerging interdisciplinary field of “network physiology”
It might seem obvious to say that everything in the human body is connected. Without a doubt, your various organs – heart, liver, lungs – work together to keep you alive, and functioning as close to normally as possible. Just think how both your heartbeat and your breathing speed up if you receive a shock – or how, in a starker example, the failure of one organ can lead to a cascade of failures in other organs, sometimes resulting in death.
But just how are our organs connected? Plamen Ivanov – a physicist at Boston University and Harvard Medical School in the US – thinks he may have at least the beginnings of an answer. Having developed and expanded upon the types of analyses found in the statistical physics of complex networks, Ivanov and others believe that the fluctuating outputs of organs, commonly considered “noise” by today’s physiologists, are in fact evidence of an underlying connectivity. Studying these fluctuations, he says, could give us an entirely new window into the workings of the human body – and help us prevent things going wrong.
Ivanov has grand ambitions. He wants to draw on statistical physics to build a human atlas, or “human physiolome” – a comprehensive map of all the interactions between organs in the human body. Like the Human Genome Project, which over 13 years uncovered the genetic blueprint of humans, Ivanov believes a human physiolome will revolutionize the analytic approaches of clinical practice. “It will pose new questions that have not been posed by the natural sciences until now,” he says.
Ivanov’s work on “network physiology”, as the field is now known, first began in the mid-1990s, when he began investigating the fluctuations in heart rates of both healthy human subjects and those suffering from sleep apnoea, when breathing during sleep becomes irregular. Everyone’s heart rate fluctuates, so if you measure someone’s heart rate to be an average of, say, 60 beats per minute, the time between neighbouring pulses could be at a rate of more like 50 or 70 beats per minute.

Ivanov found that, for people with sleep apnoea, these fluctuations were random, whereas for healthy subjects the distribution of fluctuations could be described by a single function over a wide range of timescales (1996 Nature 383 323). In other words, there appeared to be an underlying temporal structure to the heart-rate fluctuations in healthy people, in the sense that the time between any two pulses is in some way related to the time between another two pulses – seconds, minutes or even hours in the past. “It turns out that these fluctuations are not noise,” says Ivanov. “They are very structured.”
Working with colleagues at Boston and other institutions, Ivanov made some further interesting observations about scaling functions and power laws, which also describe temporal structure. He found that unique power laws could describe unique physiological states – not just being awake or being asleep, but different stages of sleep, including light sleep, deep sleep and rapid-eye-movement (REM) sleep. Intriguingly, an “asleep” power law could persist if a person was actually awake at an unusual time, for instance if he or she had travelled to a different time zone. Even more curious was that the researchers found a similarity between the diminished heart-rate fluctuations in the “half awake” physiological states of early morning and diminished fluctuations in those suffering from heart problems. The result, which goes against the popular idea that poorly hearts ought to exhibit more irregularities, could point to an underlying mechanism to explain why most heart attacks happen in the morning (2007 IEEE Eng. Med. Biol. Mag. 26 33).
The human physiolome
The application of statistical physics to physiology does not end with heart-rate analysis. Ivanov and colleagues have also found power laws to describe fluctuations in breathing, fluctuations in the movement of a person’s hands and fluctuations in a person’s gait. But the power laws appeared to extend beyond individual limbs and organs. Even in his mid-1990s work on heart-rate fluctuations, Ivanov knew that the cardiovascular and respiratory systems must be in some way “talking” to each other – after all, there was a heart-rate power law for healthy subjects, but not for those with a respiratory disorder, sleep apnoea. Indeed, in 2000 Ivanov and colleagues showed that sleep apnoea could be diagnosed not from respiratory recordings – the traditional method, which requires hospitalization – but from inexpensive, home-monitored heart-rate recordings (2000 Comput. Cardiol. 27 753). The group recently uncovered several further, independent couplings between the cardiovascular and respiratory systems, which undergo dramatic transitions from one sleep stage to another (2012 PNAS 109 10181).
Could this type of communication be continuously mapped somehow, not just between the cardiovascular and respiratory systems, but among all the body’s organs? Certainly, recent decades have seen a dramatic upsurge in the application of statistical-physics techniques to complex networks. By analysing widely available information such as Internet traffic and mobile-phone data, physicists have found new insights into underlying sociological behaviour. But whereas mobile phones and IP addresses are all similar types of node, each type of human organ is very different. Worse, the links between organs are continuously changing, and operate on a vast hierarchy of timescales.
In 2012 Ivanov and an interdisciplinary team of scientists made some headway by coming up with a concept called time-delay stability. This concept relies on measuring the time between fluctuations in the output signals of one physiological system, such as cardiovascular, and the emergence of corresponding modulations in another, such as the respiratory. According to the researchers, the longer the period during which this delay is constant – for example, the constant period during which a fluctuation in heart rate is followed by a fluctuation in breathing a few seconds later – the stronger the coupling between the two systems.
To test the time-delay stability concept, Ivanov and colleagues analysed existing sleep data taken from 36 healthy volunteers. The data consisted of heart rates, breathing and brain activity, as well as eye, leg and chin movements. The researchers found that all of these physiological systems appeared to be poorly coupled during deep sleep, but more connected when a subject transitioned into REM sleep. The links became stronger in the light-sleep phase, and stronger still when the subjects finally awoke. Based on these promising findings, it was these researchers who proposed the creation of the field of network physiology (2012 Nature Comms 3 702).
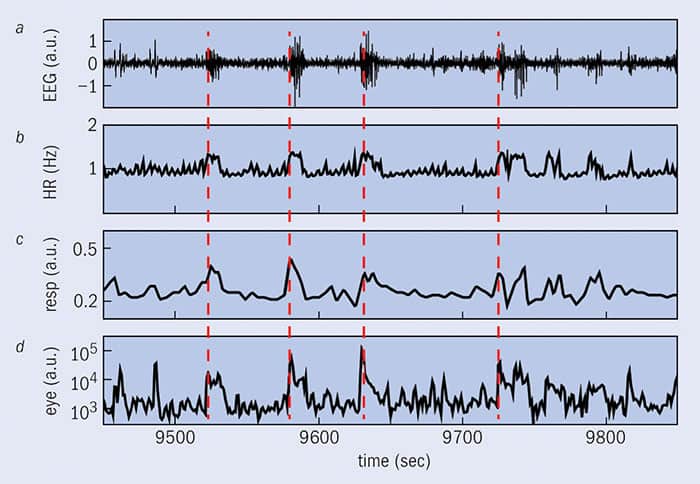
In November 2015 Ivanov and colleagues finished a more in-depth analysis of the data, and found that each organ has its own network of interactions with different areas of the brain, and with other organs (PLOS ONE 10.1371/journal.pone.0142143). Interestingly, there appear to be rules governing the reorganization of brain–organ and organ–organ interactions when a subject undergoes a transition to a different sleep stage; despite the uniqueness of the networks for each organ, these reorganization rules appeared to be the same for each of them (figure 1).
“Remarkably, these networked communications are so flexible that they can change and adjust within seconds [during] a transition from one physiological state to another – something we had not expected,” says Ivanov. “This shows an amazing flexibility and responsiveness in the way organ systems optimize and coordinate to generate different physiological functions during different physiological states.”
For Ivanov, this work could mark the beginnings of a human physiolome. And indeed he now has support: in July 2015, he was awarded a $1m (£660,000) grant by the W M Keck Foundation to develop network physiology. But he insists the path ahead will not be easy. “These are only the first steps in this new field,” he says. “Network physiology poses many new questions and challenges for which we do not yet have the necessary analytic [instrumentation] and theoretical framework.”
Even so, he continues, the challenges will be worth it to generate a new type of “big data”. The human physiolome will contain “streams of continuously recorded, high frequency, synchronized physiological signals under different physiological states and clinical conditions, [which] will change the way medicine operates today and will integrate more and more data-driven analytic approaches in clinical practice”, he says. “In the future, this new big data will have a similar impact on science, medical practice and health care as the Human Genome Project has today.”
• New Journal of Physics, from IOP Publishing – which also publishes Physics World – recently released a collection of articles in its “Focus on Network Physiology and Network Medicine”.
- Enjoy the rest of the February 2016 issue of Physics World in our digital magazine or via the Physics World app for any iOS or Android smartphone or tablet. Membership of the Institute of Physics required