A new technique to make highly complex 3D nanostructures by assembling together synthetic DNA “bricks” has been developed by researchers at Harvard University in the US. The bricks, which are like tiny pieces of LEGO, can be assembled into a wide variety of shapes and configurations, meaning that they can be used to build elaborately designed nanostructures. The resulting structures might find use in a wide variety of applications, including smart medical devices for targeted drug delivery in the body, programmable imaging probes and even in the manufacture of speedier and more powerful computer-chip circuits.
DNA nanotechnology has now been around for nearly 30 years, but it really took off with the advent of a technique called DNA “origami”. This technique, named after the ancient Japanese art of paper folding and first developed in 2006 by Paul Rothemund at the California Institute of Technology, involves folding long strands of DNA into a wide range of predetermined shapes. The resulting nanostructures can be used as scaffolding or as miniature circuit boards for precisely assembling components such as carbon nanotubes and nanowires.
Powerful though it is for making both 2D and 3D shapes, DNA origami has its limitations. To fold the DNA, several hundred “staples” must be added to the regions surrounding the single DNA strands, and each type of new nanostructure desired requires a new set of staples. Moreover, the DNA structures tend to arrange themselves randomly onto a substrate surface, which makes it difficult to integrate them into electronic circuits afterwards.
Building bricks
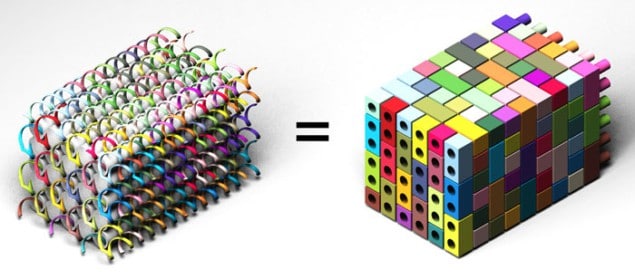
A team led by Peng Yin at Harvard first put forward its DNA-brick self-assembly technique earlier this year. Rather than starting with long DNA strands, the researchers succeeded in interlocking short, synthetic strands of DNA together to make larger structures. In fact, they managed to arrange the short strands into a “molecular canvas” by controlling the local interactions between the strands. The technique, like any DNA self-assembly method, works by exploiting the fact that the four base pairs in DNA – adenosine, thymine, cytosine and guanine – are naturally programmed to join up in specific ways: A only binds to T, while C only binds to G. So, the team was able to fabricate a collection of 2D structures using its technique by stacking one DNA brick that was 42 bases long upon another brick.
3D shapes
Now, Yin and colleagues have extended their technique to 3D. The researchers begin with an even smaller DNA-brick strand – only 32 bases long – that contains four regions than can bind to four neighbouring DNA-brick strands. The bricks are connected through 90° and so can be built out in all three directions – up, down, and out – to create a solid “master” DNA molecular-canvas cube containing hundreds of bricks. Compared with hand-assembled LEGO structures, each DNA structure self-assembles thanks to the fact that every brick is encoded with an individual sequence that determines its final position in the nanostructure. Each sequence will only be attracted to one other complementary sequence, which means that specific shapes can be created through the selection of different sequences.
The biggest advantage of the new DNA-brick technique is that any number of structures can effortlessly be made from the same master cube by simply selecting subsets of specific DNA bricks, according to the team. “We have already made more than 100 different shapes in this way (with some containing intricate cavities, surface features and channels), all of which are more complex than any 3D DNA structure constructed in the last decade. What is more, additional DNA bricks can be added, removed or modified independently without affecting other parts of the structure,” says Yin.
Complex structures
The researchers claim that the complex structures that can be made using their DNA-brick assembly technique will help advance existing DNA nanotechnology applications. “We can for example, arrange technologically relevant guest molecules into functional devices that might serve as programmable molecular probes, instruments for biological imaging and drug-delivery vehicles,” Yin tells physicsworld.com. “The structures can also be used to fabricate high-throughput complex inorganic devices for electronics and photonics applications.”
The DNA-brick structures are also entirely synthetic, whereas DNA origami is half biological. This expands the range of potential applications even further, Yin adds. “For instance, by using synthetic polymers rather than the natural form of DNA, we might be able to create functional structures that are stable in a wider variety of different environments.” The team is now busy improving its brick technique by looking more closely at DNA structure and sequence design, enzymatic synthesis for higher-quality strands and optimizing processing conditions. “We would also like to better understand the kinetic pathways involved in DNA assembly,” says Yin.
The work is published in Science.