When surgery is performed to remove cancerous tissue, one question always lingers: “did the surgeon get everything?”.
In the case of brain tumour resection, the answer is often “no”. Residual cancerous tissue at the edges of a cavity where a malignant mass has been removed can visually resemble healthy tissue and be overlooked, or may be microscopic in size.
A new tool for neurosurgeons, designed for fast and accurate detection of microscopic brain tumour infiltration in unprocessed tissue samples from surgical margins, may lead to a new era of success for brain cancer surgery.
The developers of the new FastGlioma tool, at the University of Michigan and the University of California, San Francisco (UCSF), explain that it can predict if and the extent to which glioma remains in the brain while the surgical procedure is underway. FastGlioma also provides visual heat-map guidance of the location(s) requiring additional reaction for safe maximal tumour removal.
FastGlioma combines rapid, easy-to-use stimulated Raman histology (SRH) optical imaging with open-source visual foundation models (artificial intelligence models trained on massive, diverse datasets that can be adapted for a wide range of tasks) to perform a 10 s analysis of fresh tissue specimens in operating room suites. FastGlioma proved not only significantly faster and cheaper than conventional standard-of-care MRI- and fluorescence-based surgical guidance, but in head-to-head comparisons, it significantly outperformed them in detection of two types of glioma (IDH wild-type and IDH-mutant diffuse gliomas).
Training and validation
In a prospective multicentre clinical study, principal investigators Todd Hollon of the University of Michigan and Shawn Hervey-Jumper from UCSF and co-researchers trained and validated FastGlioma to detect microscopic tumour infiltration in an international cohort of patients. They explain that “foundation modelling had not been previously investigated in studies on the clinical applications of SRH”, adding that they focused on tumour infiltration “as the most clinically important and ubiquitous problem in cancer surgery”.
The researchers trained FastGlioma using 11,462 whole-slide SRH images, divided into around four million unique 300×300 pixel SRH patches, acquired from 2799 patients undergoing surgery for suspected central nervous system tumours and/or epilepsy. They validated the model using a dataset of 3560 whole-slide images (852,000 patches) from 896 patients. Diagnostic classes of the dataset included normal brain, high-grade glioma, low-grade glioma, meningioma, pituitary adenoma, schwannoma and metastatic tumour. A subset of these had tumour infiltration categorized, ranging from normal brain to dense infiltration.
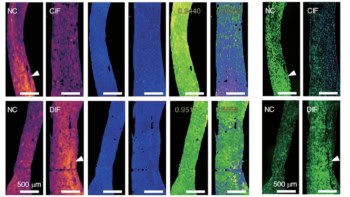
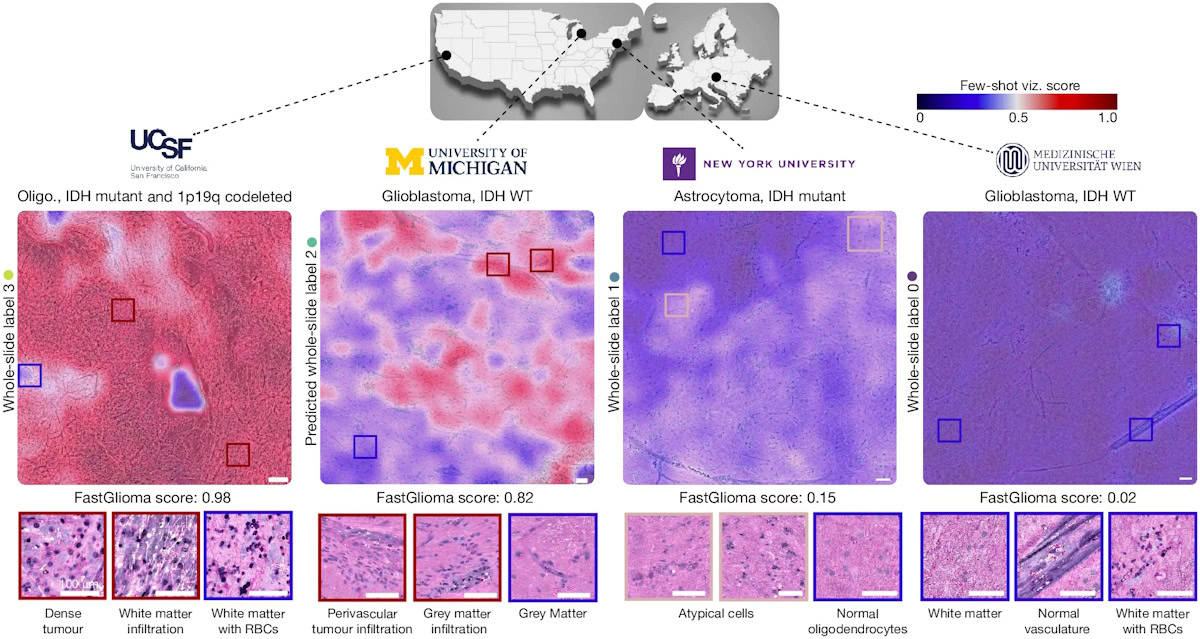
The researchers also developed a rapid visualization strategy, called few-shot visualizations. Based on FastGlioma’s self-supervised training, few-shot visualizations use a small support set of physician-selected SRH patch examples, representing a diverse selection of diffuse gliomas and normal brain parenchyma. By comparing feature similarity between the support set and the tissue sample being analysed, FastGlioma creates both a tumour-infiltration score and infiltration heat maps.
Prospective clinical testing
To test the fine-tuned FastGlioma model, three medical centres – UCSF, NYU Langone in New York City and the Medical University of Vienna – enrolled 220 patients with suspected diffuse gliomas who underwent tumour resection.
FastGlioma could detect and quantify the degree of tumour infiltration with an average accuracy of 92.1%. The tool maintained accurate tumour-infiltration scores despite significant cytological and histoarchitectural differences related to tumour grade, molecular genetics, treatment effect or WHO subtypes.
The primary end point for the study, reported in Nature, was to validate the accuracy and reproducibility of FastGlioma across various patient populations, demographics, medical centres and World Health Organization (WHO) diffuse glioma molecular subgroups. Additionally, the team aimed to compare the performance of FastGlioma with standard-of-care methods for intraoperative tumour-infiltration detection during brain tumour surgery.
To achieve this, the researchers evaluated FastGlioma as a surgical adjunct in a subset of 129 patients. Neurosurgeons sampled surgical margins at their discretion. Following SHR-imaging during the surgical procedure, the resected specimens were preserved for subsequent microscopic analysis. Expert neuropathologists scored each SRH image postoperatively to provide ground truth tumour-infiltration scores. FastGlioma significantly outperformed conventional methods, with only a 3.8% tumour miss rate, compared with a 24% miss rate for current standard-of-care surgical guidance methods.

AI tool accelerates tumour classification during brain surgery
Another benefit is that the analytic speed of FastGlioma provides a rapid and scalable alternative to conventional intraoperative pathology methods. The researchers point out that visual foundation models like FastGlioma also minimize reliance on radiographic features, contrast enhancement or extrinsic fluorescent labels to optimize the extent of resection.
The researchers also note that FastGlioma can accurately detect residual tumour for several non-glioma brain tumours, including paediatric brain tumours. “FastGlioma represents the transformative potential of medical foundation models to unlock the role of artificial intelligence in care of patients with cancer,” they write.
Future research will focus on applying a similar workflow to other human cancers, including lung, prostate, head-and-neck and breast cancer.