Protein structures found naturally in bacteria can be used as electron-microscope-compatible gene reporters in animal cells. Researchers in Germany have shown that enzymes carried within cage-like encapsulin nanocompartments (ENs) can concentrate iron, forming features that are highly visible on conventional transmission electron microscope (TEM) images. By engineering animal cells to express ENs with different sizes, the structures can fulfil for electron microscopy the role that fluorescent gene-reporter proteins have long performed in optical microscopy (ACS Nano 10.1021/acsnano.9b03140).
Fluorescent proteins are used routinely to indicate gene expression in animals and cell cultures. In this technique, the production of a fluorescent protein is made dependent upon the regulation of a gene of interest, so that when the targeted genes are expressed, so too are the fluorescent proteins.
Optical microscopes can detect the emission (typically red or green) from fluorescent proteins, but lack the resolution to discern subcellular features or the fine details of cellular networks. TEMs are usually the method of choice to study processes at this scale, but they are blind to fluorescence signals. And, until now, alternative genetically expressed, multi-channel TEM-visible labels have been lacking.
Bacterial solution
As the opacity of a sample under an electron microscope is governed by its atomic number, Gil Westmeyer, at the Technical University of Munich and Helmholtz Zentrum München, and colleagues knew that a TEM-compatible gene reporter would have to be based on the concentration of large quantities of metal. Iron, for its biological ubiquity, was the obvious choice.
Animal cells do already have the ability to concentrate iron, but not to the degree that would show up against the metal stains that are commonly used to prepare samples for electron microscopy. “Mammalian cells use ferritin as iron storage proteins, which are difficult to detect as they only have a small iron core,” says Westmeyer. In any case, an ideal gene reporter would not occur naturally in the organism, so that its expression is a distinct indicator of gene activity.
A better option is to use the ENs expressed by certain bacteria, which can accumulate much larger quantities of iron. These spherical protein structures carry an enzyme that oxidizes iron from the intracellular fluid, converting it to an insoluble form and trapping it on the inner surface of the cage.
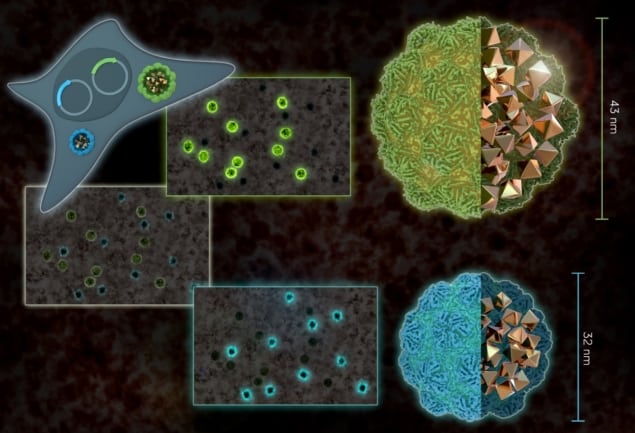
Making sure that the proteins would still express and assemble at high abundances without causing toxicity, Westmeyer and colleagues engineered animal cells to produce ENs from two species of bacteria — Quasibacillus thermotolerans (Qt) and Myxococcus xanthus (Mx). Each species forms a characteristic nanostructure with a specific size, so the researchers were able to derive a pair of reporter proteins that can be distinguished when used to simultaneously indicate the expression of two separate genes.
“Differentiating ENs by these geometrical features is possible because of their defined icosahedral geometries that make the Qt variant precisely 43 nanometres, and the Mx variant 32 nanometres,” says Westmeyer. “In analogy to the different colours of fluorescent proteins, the small ENs could be regarded as green fluorescent protein, and the large ones, red fluorescent protein, so two distinct labels for a specific cell type or cell state.”
The geometries of the two EN structures are consistent enough to allow them to be distinguished reliably by an automatic, non-human observer. When the researchers trained an artificial neural network to identify the two types of reporter protein, the system achieved better-than-human performance, suggesting the possibility of a high-throughput method to measure gene expression with little human involvement.

Getting a magnetic handle on mammalian cells
Although Westmeyer and colleagues have demonstrated only two EN reporter structures so far, other variants undoubtedly exist, which would expand the “colour palette” available to TEM studies of gene expression. Engineering the nanocages to assemble into groups, and changing how the ENs distribute themselves within cells, would add another dimension in terms of reporter differentiation.
What investigations might benefit from this new technique? One near-term application, Westmeyer suggests, is in the study of neural circuits. “Just as the Brainbow mouse can differentiate different neurons using different colours, geometrically multiplexed ENs could differentiate several neurons by their genetic identity or cellular state. The resolution of electron microscopy would allow for tracing out (segmenting) individual neurons and their synaptic connections such that a connectome can be obtained. Fluorescence microscopy does not provide an adequate resolution to achieve this.”



