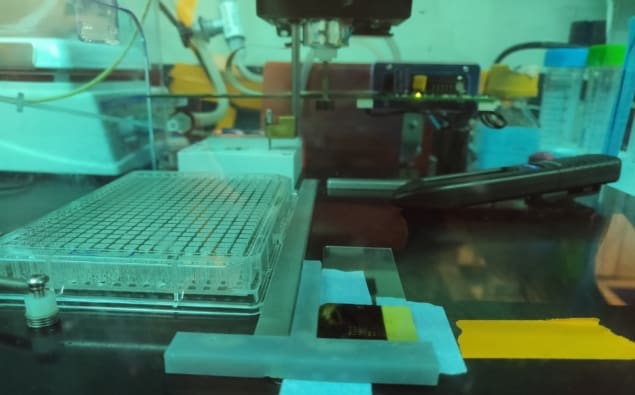
A US research collaboration has successfully adapted an ArrayIt Spotbot, usually used to monitor protein activity, to print bacteria. In doing so, the researchers have delivered an experimental platform for testing computational models of communal microbial behaviour on unprecedented small scales (Biomed. Phys. Eng. Express 4 055010).
Bioprinting is a type of 3D printing that uses cells or other biomaterials instead of ink to create biological structures. William Hynes, from SUNY Polytechnic Institute and Lawrence Livermore National Laboratory, and colleagues used the protein microarray machine to position small bacterial colonies in growth medium with high precision.
Traditional cell culture methods have not thus far provided the necessary control of the spatial distribution of cells to test predictive models of microbial interactions. This has hindered the study of communication between bacterial colonies, which underpins behaviours such as biofilm development and diffusion-based metabolite sharing.
The researchers used their novel experimental set-up to validate the computational predictions of a bacterial growth model, Computation of Microbial Ecosystems in Time and Space (COMETS). COMETS estimates the biomass of a bacterial colony as a function of time by balancing the internal cell metabolism with the external flux of metabolites. It has been tested experimentally on the macro scale, but not on the small scale used in this study.
The team used two strains of bacteria, Salmonella enterica (S. Enterica) and an Escherichia coli (E. Coli) mutant, that depend upon one another for the production of metabolites in a growth medium devoid of specific chemicals. In a control experiment, the individual strains showed little or no growth in isolation.
Each colony was made to express a fluorescent protein. The researchers showed that microbial biomass was approximately proportional to the fluorescence intensity in the imaging volume. They used this as a means of measuring bacterial biomass in vitro.
Hynes and colleagues first used the COMETS software to predict the growth of microbial biomass as a function of the separation of the two partner colonies, with colony separation varying from 1–3.5 mm. Such separations are much smaller than those used in experiments prior to this work. The research team reported good qualitative agreement between the simulation and experiment in this scenario.
The group then investigated a more complex “eclipse” scenario, in which a competitor bacterial colony is placed between the two partner colonies. The competitor was chosen to compete directly with either one of the partner colonies or with both. The team then varied the position of the competitor colony to generate different eclipse scenarios for validation.
In these more complicated scenarios, the team reported encouraging results with respect to the general trends of in silico modelling versus in vitro experiments. However, the data possess some notable disparity, including a consistent overshoot in simulations of the relative growth of E. coli, in most scenarios.
This work highlights the importance of seeking a means of validating computational models of bacterial growth across multiple length scales. It also demonstrated that bioprinting could provide a means of achieving this.
The versatility of the ArrayIt Spotbot is not limited to tightly controlling the positioning of small bacterial colonies on growth media. “For my work in developing a diagnostic assay to detect Lyme disease, the robotic pin spotter (Spotbot) is useful for making highly multiplexed protein microarrays on different surfaces,” says Eunice Chou, a PhD student working under Nathaniel Cady at SUNY Polytechnic Institute. “Here, I am using a gold-coated silicon chip, which I spot with various proteins present on the surface of Lyme disease bacteria.”