Using a combination of methods, including X-ray crystallography, small-angle scattering and live-cell imaging, an international research team has probed the structure of a potent new antibacterial agent: PyoS5. By analysing its biophysical and biochemical properties, the scientists – from the University of Oxford, Heinrich Heine University Düsseldorf, the University of Glasgow and the University of Strasbourg – mapped the path by which PyoS5 can be transported into a bacterium (mBio 10.1128/mBio.03230-19).
There is a pressing need for new antibiotics against bacterial pathogens, particularly Gram-negative bacteria, which are responsible for severe acute and chronic infections in humans and are capable of surviving under a variety of environmental conditions. For example, the World Health Organization has classified P. aeruginosa, a Gram-negative, rod-shaped bacterium, a priority pathogen due to its insensitivity to many currently used antibiotics.
Antagonism in bacteria, in which one species kills another, occurs via several routes, the most common being bacteriocin secretion. Bacteriocins are toxins, ranging from small peptides to larger protein molecules, that a bacterium produces to inhibit the growth of competing strain(s). Researchers are currently testing bacteriocins to assess their potential application as antibiotics against multidrug resistant bacteria. One of these, Pyocin S5 (PyoS5), has demonstrated great success in eradicating P. aeruginosa in animal infection models. However, the import mechanism of the PyoS5 is poorly understood.

In order to investigate a common import framework, a team led by Colin Kleanthous at the University of Oxford engineered an E. coli bacterium susceptible to PyoS5, as well as other pyocins (such as PyoS4), and proposed a generic pathway taken by these bacteriocins across the outer membrane of Gram-negative bacteria.
The process by which the PyoS5 enters the bacterium can be visualized using the analogy of a ‘fishing pole’. Three components are involved in this import process: CPA, FptA and TonB1. CPA is a surface antigen that, along with the transporter FptA, is located in the outer membrane, whilst TonB1 is present in the inner membrane. PyoS5 first accumulates on the cell surface by binding to CPA. Using its one of the unstructured ends (the N-terminus), elongated PyoS5 then contacts FptA, whilst its associated unstructured region binds TonB1. Both FptA and TonB1 drive the import of bacteriocin across the cell membrane. In addition, the team showed for the first time that PyoS4 also exploits TonB1 for its import.
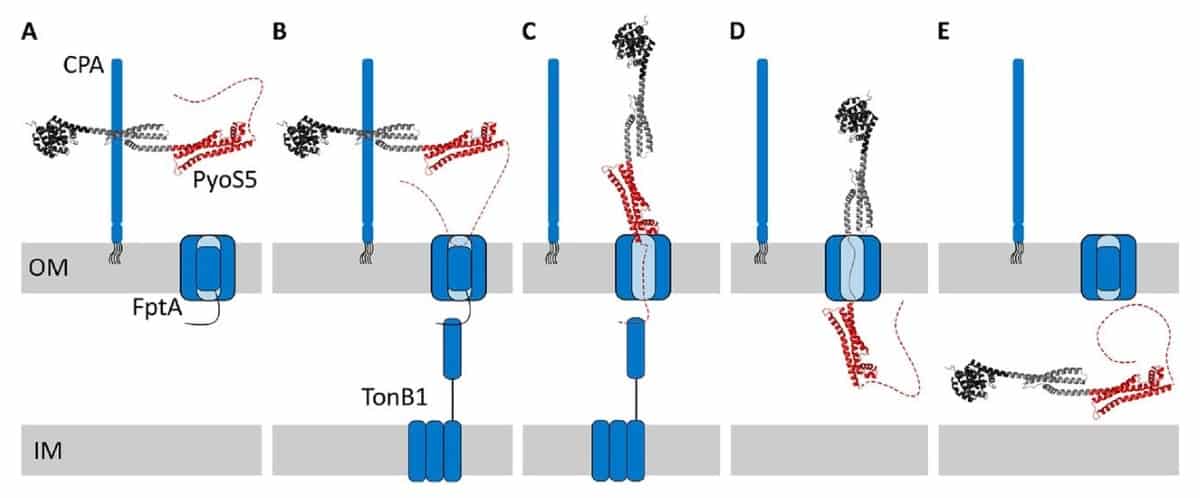
Universal uptake mechanism
As common principles are beginning to emerge, the researchers believe that these generic import mechanisms will help us to better understand other Gram-negative bacteria that can be destroyed by bacteriocins. For instance, the toxin entry mechanism in two other Gram-negative bacteria families (Enterobacteriales and Pseudomonadales) was long thought to be unrelated. However, these new findings suggest that the underlying mechanism by which some bacteriocins cross the outer membranes are fundamentally the same for both families.
“We think some of these insights likely apply to the whole family of antibiotics, with more than 3000 members,” concludes Hannah Behrens, lead author of the study.