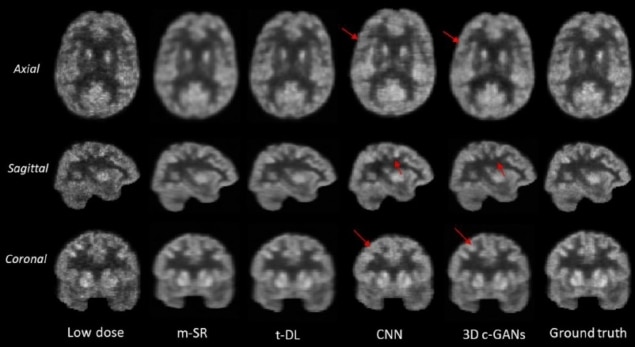
For individuals receiving multiple PET scans, especially children, there is a particular incentive to reduce the doses they receive to minimize the long-term risk of radiation-induced cancers. However, low-dose scans lack diagnostic power due to higher levels of noise. An international collaboration is using deep neural networks as a potential solution to the problem.
“Our technique uses unique machine learning algorithms – known as 3D conditional generative adversarial networks (or 3D c-GANs) – to estimate the high-quality full-dose PET images from low-dose ones,” said co-author Luping Zhou from the University of Sydney. Developed by Zhou, first author Yan Wang from Sichuan University, and co-authors in China, the US, South Korea and Australia, the new technique performed well against other methods used to synthesize full-dose PET images (NeuroImage 174 550).
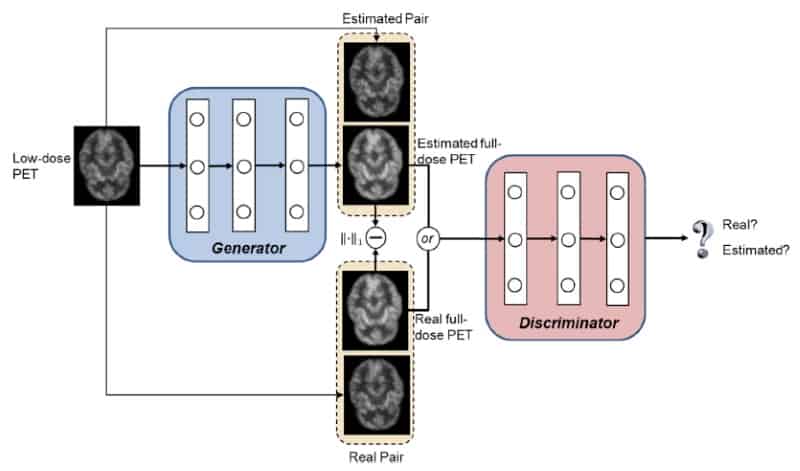
GAN models use two deep neural networks, a generator and a discriminator, which achieve the best possible result by competing against one another. In the new application, the generator’s goal is to synthesize a full-dose PET image of sufficiently high quality to convince the discriminator that the image is genuine. The discriminator’s goal is to spot that the output of the generator is not a true full-dose image.
Each network is trained using a database of pairs of low- and full-dose PET images from the same individuals. Once trained, a new individual’s low-dose PET image is then fed into the generator for it to synthesize the corresponding full-dose, higher quality PET image.
In a key feature, the technique handles 3D image data sets. Many other, previously reported techniques handle 2D axial slices independently which, though less data intensive, leads to the loss of information in the coronal and sagittal planes.
To train and validate the 3D c-GANS technique, the researchers acquired 18F-FDG brain scans of eight individuals with normal uptake and eight with mild cognitive impairment (MCI). Conventional full-dose clinical scans, delivering an effective dose of 3.86 mSv, acquired counts for 12 minutes. They were immediately followed by three-minute acquisitions that were used as low-dose scans in the 3D c-GANS model.
Following the common “leave-one-out” cross-validation approach, the researchers used data from 15 individuals to train the model and data from the remaining individual to test the model’s capabilities. The process was repeated such that the model was tested using data from all 16 individuals.
To maximize the training data set, 125 individual sub-volumes were extracted from each scan, making 1875 training samples and 125 test samples for each leave-one-out case. The final entire synthesized full-dose PET volumes were then constructed by merging the individually synthesized sub-volumes generated by the model. By maximizing the number of training samples, the likelihood of overfitting by the model was minimized, improving its potential performance in a wider clinical population.
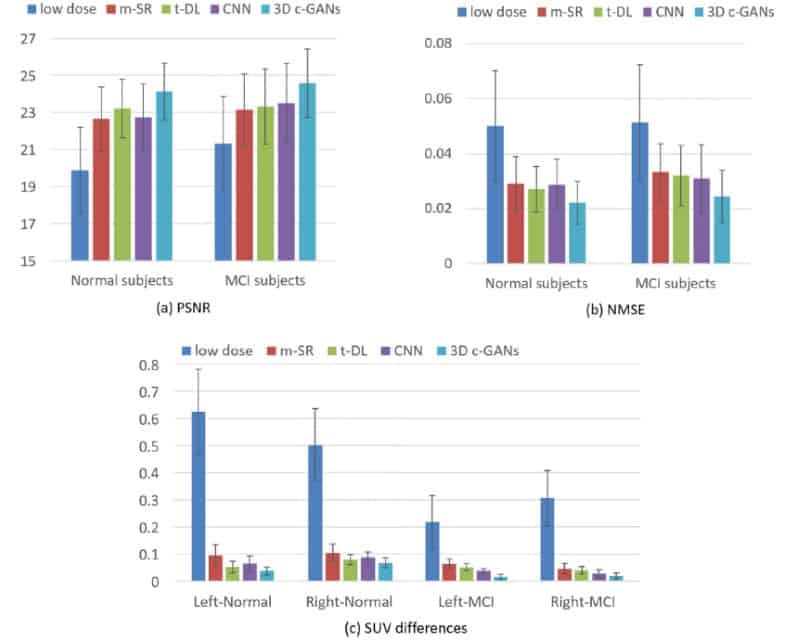
The researchers found the image quality achieved with 3D c-GANS compared favourably against three existing methods used to synthesize full-dose PET images. In a quantitative comparison, for example, the normalized mean square error (NMSE) – a measure of the difference in voxel intensities between synthesized and true full-dose PET images – was lowest using the 3D c-GANS technique. This was seen both in individuals with normal scans and those with MCI. Standard deviations in the parameter did, however, overlap between the different methods. The new technique performed similarly well when standardized uptake values (SUVs) and peak signal-to-noise ratios were examined and in a further, qualitative comparison.
Amongst several lines of further research, the authors plan to investigate a multi-modality approach to full-dose PET synthesis, incorporating clinical CT or MRI scans that are acquired alongside the PET scans. They also plan to increase their training database size to improve 3D c-GANS generalizability – its ability to work effectively in the clinical patient population at large.



