Curved lipid membranes are abundant in cells. Most folded proteins are thought to interact with the membrane directly via specialized folded regions, which prefer curved regions. These proteins contain large, disordered domains (independent three-dimensional structures inside the protein) without any well-defined structure. Indeed, these disordered regions resemble random coils, tangled like cooked spaghetti, yet they can still sense and localize to curved membrane regions. As a result, it has not been clear how proteins sense curved membranes to localize there.
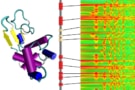
To understand the interactions between disordered protein structures and curved regions of the membrane, researchers from the University of Texas led by Jeanne Stachowiak tethered small unilamellar vesicles (SUVs) to a glass surface and tagged them with a red fluorescent dye. The researchers then incubated the SUVs with a disordered region of a protein that plays a role in sensing natural membrane deformation. By truncating the region from the rest of the protein, and tagging it with a green dye, the researchers were able to quantitatively measure how much protein had accumulated on the red SUVs as a function of SUV diameter and chain length.
Observing the mixture using fluorescence microscopy, the researchers found that the fluorescence intensity of the protein (green) increased as the SUV diameter decreased, which they could see from the lower red fluorescence coming from the labelled SUVs – “smaller SUVs appeared more green while larger SUVs appeared more red” reported the researchers. When the disordered region is longer, the length difference between the protein region and SUV diameter increases, which suggests a stronger curvature sensing when the protein is longer.
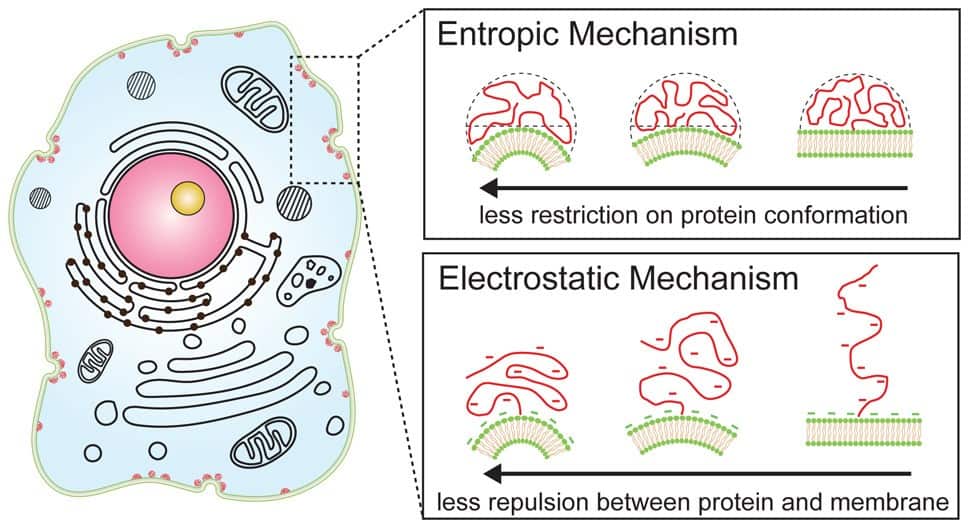
The effects of disordered protein region length suggest the role of entropy in curvature sensing since there are more possible configurations for a longer region than a shorter one. Simply put, the disordered protein is merely maximizing its entropy as a curved membrane allows the coiled chain to occupy more volume with more possible configurations than a flat one.
Ionic strength dictates which force drives curvature sensing
The researchers then modified the salt concentration of their solution to change the ionic strength felt by their disordered domains. At high salt concentrations, the longer chains were more sensitive to membrane curvature. Conversely, in the low salt regime, they observed no difference in curvature sensitivity for different chain lengths. However, in this regime, the curvature sensing of the shorter chains was much stronger than at high salt concentrations.
Amino acid orchestra trains machine learning algorithms to design proteins
The effects of salt concentration suggest that for smaller disordered chains, electrostatic forces drive membrane curvature sensing behaviour as curved membranes allow for increased distance between repulsive amino acid chains and charged membrane lipids. Adding small amounts of an anionic lipid (DOPS) into their SUVs increased curvature sensing for all cases, with the strongest effect being observed for the shortest disordered chain, reinforcing the case for electrostatic forces dominating curvature sensing for short chains.
The researchers’ report in the Journal of the American Chemical Society, shows how non-specific interactions can play important roles in very specific interactions – such as that of proteins and cell membranes. Their results remind us that, despite the complexity of biology, simple physics processes still play important roles in nature.