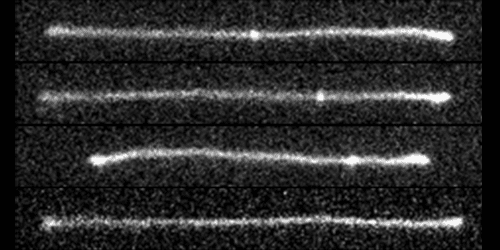
Knots have been removed from DNA by stretching the molecular strands with an electric field. The work was done by researchers in the US, who have quantified how stretching causes a knot to move along DNA so that it vanishes when it reaches the end of the strand. The experiment also gives insights into how nanoscale knots can become jammed on a strand. Understanding how to untangle knots in molecular strands could improve DNA sequencing technologies and lead to better polymer-based industrial processes.
Ropes, earphone cables, necklaces and other strands can quickly become tangled into knots – a frustration of daily life that has fascinated physicists. The longer a strand, the more likely it is to get tangled – and the same principle applies to long chain molecules (polymers) including DNA.
Scientists are keen to understand knots on a molecular scale because tangling affects industrial polymers as well as biological processes involving DNA interactions. And the ability to unpick knots could aid a range of industrial and biomedical applications.
Knotty blockage
In biomedicine, for example, there is a push to achieve rapid sequencing of an individual’s genetic make-up and this is driving scientists to develop new techniques that read longer and longer sections of DNA. One promising method involves threading DNA through a nanochannel, but this channel would be blocked by knots in the DNA.
The motion of knots along molecular strands has been studied in great detail using computer simulations, however there have not been many experimental studies to date. A few experiments have detected knot movement, but quantifying knot mobility at the nanoscale has proved challenging. Now Patrick Doyle and colleagues at the Massachusetts Institute of Technology (MIT) in the US, have come up with a better way of characterizing knot mobility in the lab.
Using a special T-shaped microfluidic channel, the researchers stretch out a single DNA molecule and observe knot mobility using fluorescence microscopy. Their experiment begins by creating complex knots of different topologies in DNA by applying an alternating electric field to the strands. Then a divergent electric field is used to trap and stretch a knotted strand within the microfluidic channel.
DNA tug-of-war
The divergent electric field pulls on either end of the negatively charged phosphate backbone of DNA. “It kind of creates a tug-of-war on the DNA,” says Doyle. The electric field also drives knots towards the nearest end of a DNA strand, where they untie. The experiments confirm predictions from computer simulations that knots are able to diffuse along uniformly stretched chains.
“It’s one way to control the dynamics of knots in DNA,” says computational biophysicist Davide Marenduzzo at the University of Edinburgh, who hopes the technique can be developed to improve genomic technologies.
Computer simulations done by Doyle and colleagues suggest that knot mobility is mediated by “self-reptation”, which is a snake-like thermal motion observed in microscopic strands. The speed at which a knot diffuses along a strand is thought to be determined by a competition between deterministic and Brownian motion.
To find out how tension in the DNA affects mobility, the researchers increased the electric field, which boosts the “stretch” applied to the DNA. They found that increasing tension past a critical point slowed knot diffusion and at higher tensions, the knots began to jam.
In a jam
“This idea that you could jam molecular knots has been floated around since the 1970’s, but not really accepted because it was never experimentally proven,” says Doyle. “I think we have provided good proof now.”
Intramolecular friction between DNA atoms is thought to cause jamming, with increased tension pushing atoms into closer contact and preventing self-reptation within knots. Cristian Micheletti, professor of statistical and biological physics at the International School for Advanced Studies (SISSA) in Italy is excited about the potential of this technique. “It gives a unique opportunity to understand friction at the molecular level,” he says.
Both Micheletti and Marenduzzo are particularly eager to see an extension of this study to examine the mobility of different types of knots.
Doyle has a multitude of plans to use this microfluidic system to tease apart a number of knotty questions. But at the moment he is focused on a project for moving DNA through 20 nm holes, work that has potential to feed into the sequencing of long strands of DNA.
The research is described in Physical Review Letters.