A new mathematical model that describes how proteins self-assemble into stacks known as amyloid fibrils could aid the search for drugs to treat diseases like Alzheimer’s and Parkinson’s. The model, developed by researchers at the University of Cambridge in the UK and Lund University in Sweden, is based on rate equations and confirms that the microscopic reaction steps behind protein aggregation occur in a catalytic manner, similar to reactions involving enzymes.
The self-assembly of proteins into amyloid fibrils is a natural biological process known to be involved in several diseases, including Type II diabetes as well as Alzheimer’s and Parkinson’s. By unravelling the reactions that lead these biofilaments to proliferate, researchers hope to gain insights into how to treat such diseases. Ultimately, such knowledge could even make it possible to prevent these illnesses by developing drugs that inhibit the protein self-assembly processes at their heart.
Two-step catalytic process
Researchers know that biofilaments assemble from protein monomers in a slow “primary” nucleation-reaction process, followed by a faster process that allows the filaments to elongate. Secondary reaction processes – including nucleation of new filaments on the surface of existing ones, filament breakage and branching – frequently accompany these first two steps.
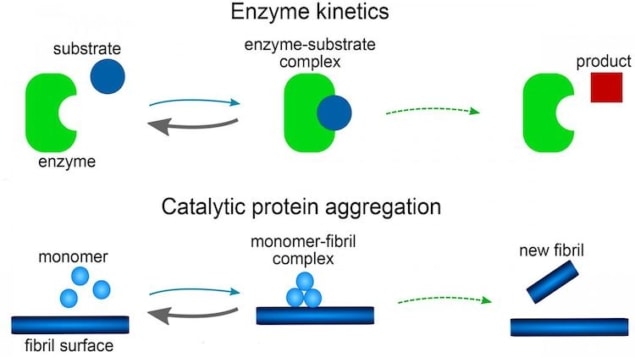
Previous work has shown that the elongation step is best described as a two-step catalytic process controlled by the so-called Michaelis-Menten rate law, which was first employed in 1913 to describe the rates of enzyme-driven reactions. The secondary nucleation step – which is known to be crucial for the aggregation of an Alzheimer’s-associated protein called Ab40 – is described by a similar rate law and is also catalytic.
Catalyzed aggregation
A team led by Tuomas Knowles and Alexander Dear has now built on these insights to develop simple, highly general mathematical equations that model the kinetics of amyloid fibril formation and changes in protein aggregate concentrations over time. The team also applied this model to experimental data on the aggregation of Ab40.
Experiments had hinted that Ab40 begins aggregating at interfaces – for example, the surface of a liquid solution or the glass wall of the test tube in which the experiments were performed. Dear says that the close match between their rate-law model and earlier in vitro results supports this hypothesis, indicating that interfaces may play an enzyme-like role in promoting protein aggregation. “The new model sheds light on the nature of aggregation by showing that the individual steps in the aggregation process (filament growth, and both primary and secondary nucleation of new filaments) are typically all catalytic in nature,” he tells Physics World. Previous models, he adds, did not take this into account.

Pulling proteins through a pore dissolves aggregates
Enzyme-like effects
A simpler form of the model also enables the researchers to see how enzyme-like “saturation” in each step affects the overall proliferation of amyloid fibrils, as the various catalytic surfaces become fully occupied at high protein concentrations. “We can thus determine the protein concentrations at which each reaction step saturates from the measured fibril growth curves,” Dear says.
The discovery that primary nucleation of Ab40 is catalysed by interfaces also has implications for the way proteins aggregate in the human body. Since such heterogenous nucleation is highly environment-dependent, Dear says that relatively small changes in the biochemical environment in the brain or body could dramatically change the propensity for amyloids to form. Ultimately, the researchers, who report their work in the Journal of Chemical Physics, believe that their result could be used to develop potent inhibitors of amyloid formation, and thus new treatments for diseases like Alzheimer’s.



