Bacterial cultures are highly diversified, with each bacterium very different to another. And just as it is not fair to tar everyone with the same brush, researchers are looking for tools with which to investigate the properties of individual cells. Indeed, studying cells one by one is extremely helpful for understanding the dynamics and behaviour of cellular populations.
Locating and studying many individual bacteria over time, however, is not an easy task. Most single-cell techniques come with an undesirable trade-off: they can measure many single cells for a very short time frame each; or track fewer cells over longer times. Selectively picking and retrieving the “odd ones out” for further analysis is even more difficult.
A recent study by Scott Luro and colleagues from Harvard University reports on a new tool that overcomes this trade-off (Nature Methods 10.1038/s41592-019-0620-7).
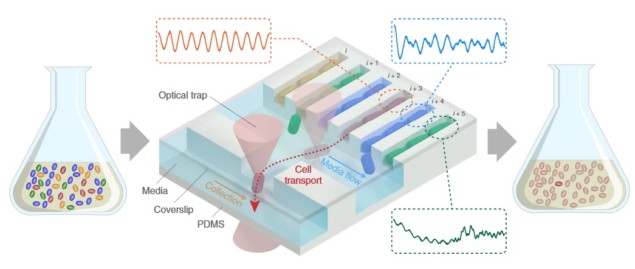
The research team used a microfluidic device, known as the mother machine, to localize thousands of individual bacteria in microscopic channels. Once a single bacterium, referred to as the mother cell, enters one of the channels, its growth is constrained to a single direction so that daughter cells can be characterized with time-lapse microscopy over many generations. Such lengthy observations capture dynamic cell-to-cell differences while providing enough data to reliably quantify bacterial traits (the so-called phenotype, such as shape, growth rate or behaviour).
After the long-term screening process, an external optical trap can pick up any bacterium of choice using a focused low-power laser beam to attract the bacterium in the laser focal spot. This allows the safe transport of cells to a second parallel collection channel. From here, single bacteria are flushed out of the chip and collected for further analysis, such as genome sequencing or plating. This process enabled the researchers to link specific observed phenotypes on chip to their genomic origins.
“The mother machine has enabled quantitative measurements and analyses of many subtle but important biological phenomena,” says Luro, lead author and graduate student in the Johan Paulsson Lab at Harvard. “The tool we created transforms this powerful imaging platform into a screening device, with the ability to cleanly collect live individual cells of interest.”
Large-scale screening of microbes
In a set of proof-of-principle experiments, the researchers created a mock microbial culture containing three labelled sub-populations mixed into a larger population of unlabelled bacteria to simulate a screening run. The three minority populations showed different phenotypes to the rest: they were fluorescent bacteria emitting red, yellow or cyan light. After growing the culture for 24 hours, the team separately picked three individual different-coloured bacteria, flushed them out of the chip and plated them to prove successful isolation of a bacterium from each sub-population.
In a second series of experiments, the team studied synthetic gene oscillators, namely systems that periodically produce certain proteins induced by deliberate and specific modifications of bacterial plasmids. An important requirement for the study was long-term imaging to accurately measure the amplitude and phase of the oscillations, for which the newly developed chip provided a perfect tool. In addition, the large size of the screened population enabled exploration of oscillatory properties from many different genetic variants. These features allowed the researchers to reliably measure and design some of the most regular periodic gene oscillators known to date.
A new tool for efficient genetic screening
The method developed by Luro and colleagues offers potential for many applications. The ability to screen such a huge number of phenotypes, pinpoint those of interest, and strongly link them to their genotype, could be the cornerstone of new genetic screening procedures. Furthermore, the device enables such screening to be performed at the single-cell level.
“We believe this screening platform could be particularly useful for generating ‘designer’ cells engineered to carry out complex functions, since live top-performing variants are physically collected. This circumvents the need for strain reconstruction, as would be required for similar methods reliant on in situ barcoding,” explains Luro. “We are also looking beyond microbes and have begun adapting our chips for mammalian cell cultivation and isolation, which looks promising.”
In turn, the method allows users to trap and track thousands of bacterial lineages and then retrieve down to a single bacterium off the chip, be it a phenotypic outlier, a specific mutant or your favourite-looking bacterium.