A new platform to image single molecules in 3D could allow researchers to investigate complex biomolecular interactions within biological cells. The platform, which uses spectroscopic single-molecule localization microscopy (sSMLM) improved with a two-mirror system, can image molecules at much greater depths than was possible before.
Spectroscopic single-molecule localization microscopy is a relatively new extension of super-resolution microscopy and it can provide information on where molecules are located in space and how these molecules interact with light. Since sSMLM records the full fluorescent spectra of emission from all the molecules in a sample, it can, in principle, differentiate unlimited numbers of molecular species within a predetermined spectral range.
The system fundamentally suffers from a limited photon budget, however, because the emitted photons have to be divided into two independent channels for spatial and spectral imaging. This means that it is challenging to localize molecules in 3D using sSMLM in its current form.
A pair of mirrors improves sSMLM
Researchers from Northwestern University led by Hao F. Zhang and Cheng Sun have now improved this platform by combining existing sSMLM with a pair of mirrors, so allowing it to image molecules in 3D at a much greater depth without inducing additional photon loss.
To extend sSMLM to 3D, the researchers initially tried adding a cylindrical lens into the spatial imaging channel, but the inherent astigmatism of the lens makes it difficult to calibrate the light spectra of different molecules and produces non-uniform lateral resolution at different depths.
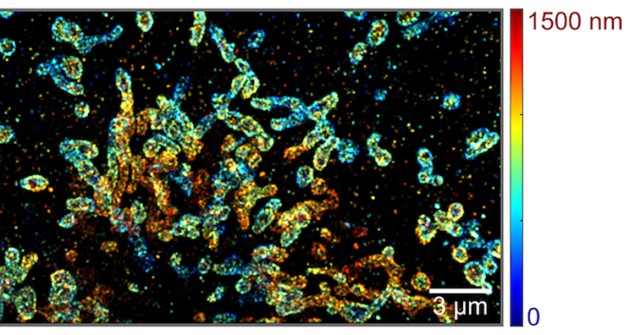
“The two mirrors work by introducing an optical path length difference between the spatial and spectral imaging channels that improves the way the system exploits photons,” explains Zhang. Unlike a lens, a mirror does not generally attenuate the light it reflects, which means that more photons can be used to create a sharper image and extend imaging into the 3D-depth range.
Detecting single molecular emission
“The optical path length difference in fact generates different focal planes for spatial and spectral imaging and we are thus able to detect single molecular light emission with different point-spread-function sizes in the spatial and spectral imaging channels,” he says. “We encode the depth information of individual molecules as the difference between the point-spread-functions and retrieve this information using a pre-determined calibration curve directly correlated with the optical path difference.”
The new system can detect the minute differences in molecular emission from every different molecule in a sample and analyse the spectra of these molecules to differentiate between them. It can be used to image molecules and biomolecules, such as DNA, RNA and proteins that can be labelled with fluorescent tags, says Zhang. Any nanoparticles that blink can also be used as an imaging agent.
Super resolution microscopy gets a dose of deep learning
A lateral spatial precision of 20 nm
Indeed, the researchers have already used their technique to image microtubules and mitochondria in COS-7 cells immuno-stained with Alexa Fluor 647 and CF 660C dyes. They showed that they could image the structures with a lateral spatial precision of 20 nm at an average photon count of 560, and a spectral precision of 4 nm at an average photon count of 1250. The spatial resolution in the vertical direction is 50 nm.
“The new platform will allow researchers to investigate complex biomolecular interactions within cells, particularly those related to transcription processes, and understand how their spatial organization impacts specific molecular biology/biochemistry mechanisms associated with diseases,” Zhang tells Physics World. “For instance, we are now characterizing how proteins are arranged in nuclear pore complexes, which are the nanochannels that transport genetic information across the nuclear envelope to the cytoplasm. Our technique will allow us to simultaneously detect multiple subunits in the nuclear pore complex in 3D.”
The researchers, reporting their work in Optica, are now trying to further push the limit of spatial resolution in their technique by looking for new way ways to correlate the spatial and spectral information obtained. “We would also like to understand the fundamental limits of this resolution,” says Zhang.



