The virus-replicating machinery of the SARS-Cov-2 virus and the ribonucleic acid (RNA) molecules it produces as it replicates are located in physically distinct areas of an infected cell. This observation, made by researchers at Stanford University in the US using a new multicolour super-resolution microscopy imaging technique, provides fresh insights into the life cycle of coronaviruses and could aid the development of new therapeutics to fight them.
The coronavirus responsible for the COVID-19 pandemic is one of seven coronaviruses that can infect humans, and the third within 20 years to cause life-threatening disease. To develop improved treatments to fight this and other such viruses, researchers need to better understand the biology of the coronavirus’ genome RNA, or gRNA, during infection, as this RNA contains all the “blueprints” required for making the proteins needed for the virus to replicate.
A team led by William Moerner and Stanley Qi at Stanford has now studied coronavirus infection at the nanoscale using high-throughput confocal and super-resolution microscopy. The researchers used a less harmful human coronavirus, HCoV-229E, as their model, because it is similar to SARS-Cov-2 in that it contains an envelope studded with protein spikes surrounding a strand of gRNA. These proteins include those that make gRNA copies and those that assemble into the “packaging” that wraps around the gRNA to make new viruses.
Differently coloured fluorescent tags
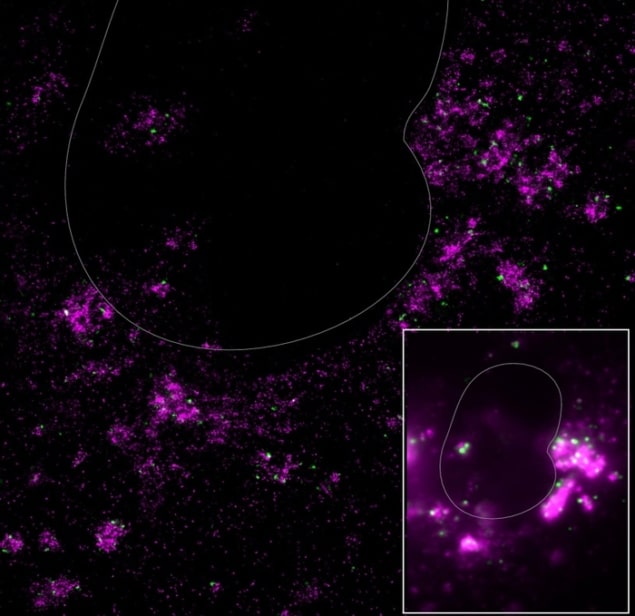
In their study, Moerner, Qi and colleagues focused on gRNA and another type of RNA, double-stranded RNA (dsRNA), which makes new copies of the virus. They began by labelling the gRNA and dsRNA with differently coloured fluorescent tags – magenta for the gRNA and green for the dsRNA – so that they could study where the two types of RNA migrated in a cell. The technique they used to do this, confocal fluorescence microscopy, is good at recording light emitted from the fluorescent labels. However, its spatial resolution is limited to imaging structures 250 nm in size or bigger. This is a problem because coronaviruses are half this size, and the proteins and RNA they contain are smaller still, at 50 nm and 10 nm or less. The result is that the images obtained are blurry and cannot distinguish between the different biological structures, let alone pinpoint where they are located in a cell.
To overcome this problem, the researchers turned to super-resolution fluorescence microscopy, which brings these structures into sharper focus, allowing objects as small as 10 nm across to be imaged. This is well below the optical diffraction limit. Using this technique, Moerner, Qi and colleagues report that they observed an “amazing galaxy” of structures, including a dark “sky” of non-overlapping bright magenta clusters and green stars.
The new observations are in stark contrast to the images obtained using confocal microscopy, which showed blurry white clouds in addition to spots of green and magenta, suggesting that the dsRNA and gRNA could be in the same areas in the cells and possibly even enveloped together. The images from the more complex multi-colour super-resolution fluorescence technique contradict this hypothesis, revealing that the different types of RNA are physically separated in the cell.
Temporary dsRNA storage centres
Further experiments confirmed that the viral replication occurs in a part of the cell known as the endoplasmic reticulum, something that researchers already knew, and that the gRNA formed then “buds off” into the cell to become packaged into a fully-formed virus. However, in contrast to previous studies, Moerner, Qi and colleagues observed that as well as being in the endoplasmic reticulum, the dsRNA is also found in large spheres that can be up to 450 nm in size. These spheres do not contain any gRNA.
Optical microscopy – how small can it go?
The researchers say that these dsRNA spheres could serve as temporary dsRNA storage centres while new viruses are “being packaged and shipped out”. While they are as yet unsure what makes the virus produce these temporary storage sites, they hope their technique could shed more light on this puzzle in the future. What is more, discovering where viral infection takes place could lead to better treatments to fight these pathogens.
“While the optical method we developed is not completely new, combining it with specific labelling of the important virus RNA components is new, to our knowledge,” Moerner tells Physics World. “The technique could be applied to many other coronaviruses, including new variants, and to monitor how they react to various drug treatments. Indeed, the Stanford team hopes to do just this in future work.”
The present study is detailed in Cell Reports Methods.



