Researchers in California have developed a deep-learning framework that performs in vivo virtual histology of intact skin. The framework rapidly transforms label-free reflectance confocal microscopy (RCM) images into virtually stained images that exhibit similar features to traditional histology of the excised tissue. The first application of virtual histology to intact, unbiopsied tissue, the study provides an important step towards the use of virtual histology technology for clinical dermatology.
“This process bypasses several standard steps typically used for diagnosis – including skin biopsy, tissue fixation, processing, sectioning and histochemical staining,” explains senior author Aydogan Ozcan from the UCLA Samueli School of Engineering in a press statement. “Images appear like biopsied, histochemically stained skin sections imaged on microscope slides.”
The current standard for diagnosing skin disease is based on invasive biopsy and histopathological evaluation. Non-invasive imaging techniques such as RCM could help prevent unnecessary skin biopsies. RCM is an emerging microscopy technology that has been available for use in clinical dermatology over the past decade. It works by detecting light backscattered from structures within the skin and provides in vivo imaging at near-histologic resolution.
Adoption of RCM, however, has not been widespread. The researchers attribute this to various factors: in vivo RCM does not show the nuclear features of skin cells seen in traditional microscopy; and the acquired images are greyscale, limiting contrast between structures. In addition, dermatologists accustomed to interpreting tissue pathology in the vertical plane may have difficulty interpreting images presented in the horizontal imaging axis of confocal microscopy. As such, analysis of RCM images is relatively challenging and requires specialized training.
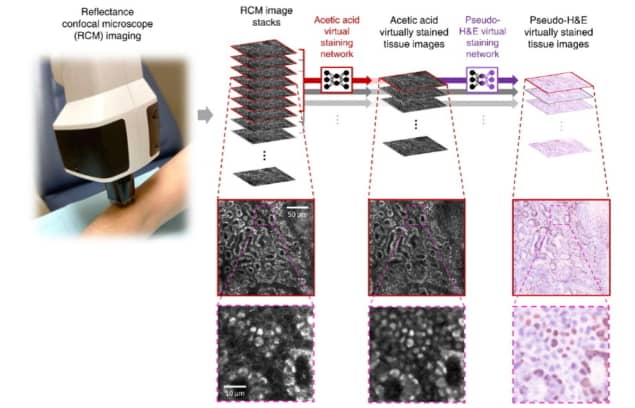
Ozcan, along with UCLA’s Philip Scumpia and Gennady Rubinstein from the Dermatology and Laser Centre, led the research initiative to virtually stain RCM images into a user-friendly format for dermatologists and pathologists. Their research team trained a convolutional neural network to rapidly transform RCM images of unstained skin into haematoxylin and eosin (H&E)-like 3D images with microscopic resolution.
To train the neural network to correctly stain nuclear features, the team devised a way to provide nuclear contrast to cells within the skin, a feature that’s normally lacking in RCM images. The network was trained under an adversarial learning scheme, which takes ex vivo RCM images of excised unstained tissue as inputs and uses microscopic images of the same tissue stained with acetic acid (to provide nuclear contrast) as the ground truth.
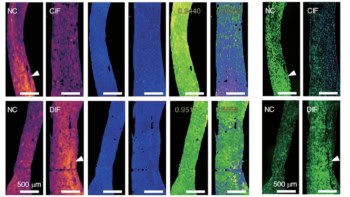
A board-certified dermatologist trained in RCM imaging and analysis used a VivaScope 1500 system to capture RCM images of 43 patients. These included three RCM mosaic scans (in which multiple images are scanned over a large area at the same depth to increase the field-of-view) and two z-stacks (through different skin layers) of both normal skin and suspicious skin lesions for each patient. In addition, discarded Mohs surgery skin tissue specimens from 36 patients with and without residual basal cell carcinoma (BCC) tumour were RCM-imaged ex vivo.
The researchers then applied virtual staining to the greyscale images to match the characteristics of H&E staining, in which cell nuclei and cytoplasm are stained blue and pink, respectively. They describe the processes in detail in Light: Science & Applications.
The final ex vivo training, validation and testing datasets used to train the deep network and perform quantitative analysis comprised 1185, 137 and 199 ex vivo RCM images of unstained skin lesions and their corresponding acetic acid-stained ground truth, obtained from 26, four and six patients, respectively.
Performance assessment
To evaluate their model, the researchers segmented virtual histology images of normal skin samples and their ground truth images to identify cell nuclei. They found that the virtual staining achieved around 80% sensitivity and 70% precision for nuclei prediction. Examining nuclear morphological features in the images revealed that parameters calculated using virtual staining matched well with those using the ground truth images.
The researchers also examined the use of both 2D and 3D image stacks as input to the deep network. They determined that using a 3D RCM image stack containing multiple adjacent slices as input produced a better virtually stained image than using a single 2D stack, which produced suboptimal blurred images.
The trained neural network was tested on new RCM images of unbiopsied BCC and pigmented melanocytic nevi, to determine whether RCM images with virtual histology could be used diagnostically. The researchers report good concordance between the virtual histology and common histologic features, in the areas of skin most commonly involved in pathological conditions. Virtually stained RCM images of BCC showed the same histological features used to diagnose BCC from skin biopsies via H&E histology. The virtual staining also successfully inferred pigmented melanocytes in benign melanocytic nevi.

Mid-IR spectrometer provides non-invasive skin cancer detection
The researchers note that further investigation is required to understand how virtual histology affects diagnostic accuracy, sensitivity and specificity, compared with the greyscale contrast of RCM imaging. They plan to collect more BCC and additional BCC-subtype data to assess their deep-learning network’s ability to detect cell nuclei inside basal cell tumour islands.
“Future studies will evaluate the utility of our approach across multiple types of skin neoplasms and other non-invasive imaging modalities toward the goal of optical biopsy enhancement for non-invasive skin diagnosis,” the researchers conclude.