Visualizing and understanding complex biological systems at the cellular level is an endeavour that requires the best efforts of both biologists and physicists, explain Paul O’Shea, Mike Somekh and William Barnes
Since the discovery of X-rays in 1895, curiosity as well as clinical need has produced huge advances in our ability to visualize structures inside the human body. X-ray radiographic imaging led the way, followed in 1957 by the use of gamma-ray cameras, and then ultrasound imaging, magnetic resonance imaging (MRI), and positron emission tomography (PET). Together, these tools, in which physics plays a key part, underpin modern clinical-imaging practice.
The length scales that these techniques interrogate, however, leave much to be desired if one is interested in the very small. Clinical MRI, for example, can only resolve structures down to 100 µm. While some living cells are more than 80 µm across, interesting and important cellular processes — such as signalling between cells — may take place over length scales of much less than 1 µm.
Living cells are essentially defined by their complex spatial structures — for example the doughnut shape of red blood cells and the elongated projections (known as axons) of neuronal cells are key to their functions. Underlying these broad morphological characteristics, however, are much finer-scale molecular assemblies, such as the cytoskeleton (a protein “scaffolding” that stabilizes the larger-scale intracellular structures) and microdomains within cell membranes, which are the locii of many molecular signalling events.
Any technique to study the properties of biological molecules and their many interactions should ideally provide spatial information, because researchers increasingly need to integrate information about the interactions that underlie a biological effect with data on where in cells these interactions take place.
Short wavelength X-rays can, of course, be used to provide information on very small length scales and can even produce images of individual molecules — as in X-ray crystallography. The downside is that such radiation can severely damage biological materials. X-rays with slightly longer wavelengths and lower energy — which can be produced by synchrotrons — are, however, much less destructive to tissue. Indeed, these “soft” X-rays can be used to generate 3D images of living cells with a resolution of up to 10 nm, as has been shown by research at the Advanced Light Source synchrotron at the Lawrence Berkeley National Laboratory in the US. Unfortunately, even soft X-rays are destructive and difficult to handle, and they require access to a synchrotron.
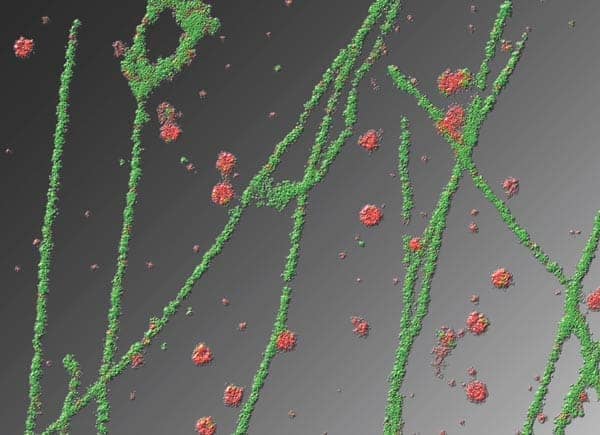
Most techniques used for cellular imaging, therefore, tend to be optical, exploiting mainly the ultraviolet and visible parts of the electromagnetic spectrum. Of these techniques, fluorescence microscopy has become the most important because it is very sensitive, enormously versatile and relatively easy to implement. Fluorescence is the phenomenon by which certain molecular structures, known as fluorophores, emit photons when excited via irradiation with light of a specific wavelength. This emission typically occurs over a timescale of 1–10 ns, which is suitable for measuring the movements and re-orientations of molecules within cells, thus allowing many biological processes to be followed.
In rare cases, the biological molecule of interest is inherently fluorescent. But usually fluorescence has to be “built into” the molecules that researchers wish to study by tagging them with a fluorophore. The tag can even be a whole protein in its own right, such as the green fluorescent protein (GFP), which is attached to and expressed at the same time as the protein of interest and only fluoresces when both are actually manufactured by the cell.
The energy, momentum, polarization state and emission time of photons emitted by fluorophores can all provide vital information about biological processes at the microscopic and nanoscopic scales. The polarization of fluorescence photons, for instance, is affected by the orientation of the fluorophore — and hence of any protein to which it is attached — and can therefore provide information about the molecular dynamics of the molecule of interest.
Much work is being done by physicists and biologists that takes advantage of each of the attributes of fluorescence — developments that are symptomatic of far-reaching changes in the way that we do science across discipline frontiers (see “Life changing physics”).
Biology meets telecoms
Whatever technique is used for imaging intracellular structures and processes, the essence of the imaging problem is the same: to extract as much information as possible from a biological sample. Some of the challenges that need to be overcome in achieving this goal, therefore, are akin to those encountered within the field of information transmission.
Communication involves sending signals — often in the form of optical pulses — down a channel to a receiver, where they are decoded and translated into “useful” information. Similarly, molecules of interest that are “lit up” in some way convey information through a microscope (the channel) to a detector or the eyes of the human investigator (the receiver), where this information reveals features of cellular structure.
In the late 1940s Claude Shannon, an engineering physicist working at Bell Labs in the US, developed some of the fundamental rules that define the capacity of an information channel. He quantified, for the first time using well-defined physical properties, the amount of information that could be transmitted down a channel, and his rules also allowed researchers to calculate the precise degree to which the data would be corrupted by imperfections and noise. These same rules apply to all communication channels, whether they convey eBay bids and Web applets across the Atlantic Ocean or information about molecular dynamics within a living cell.
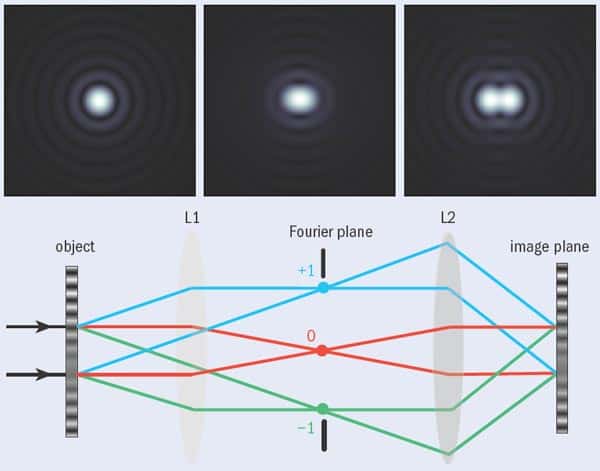
Studying living cells requires collecting enough photons to form an image. This is often far from easy — partly because cells are very small, and researchers are often interested in only a few fluorophore-labelled molecules within them. But what makes the job harder is that any beam of light of finite size undergoes diffraction and therefore spreads out. This limits the minimum diameter of the spot of light formed at the focus of a lens and provides a limit to the resolution of any traditional microscope systems. As was shown (separately) by Lord Rayleigh and Ernst Abbe over 100 years ago, this minimum diameter is approximately half the wavelength of the illuminating light (see “Criteria for success”). For conventional fluorescence microscopy that diameter is about 200 nm, meaning that features smaller than this cannot be resolved using this technique.
Microscopy therefore faces two related challenges: extracting information from as small a region as possible; and extracting as much information as possible from that small region. Doing the latter is not easy because each fluorescent molecule will only yield a finite number of photons before it stops fluorescing. It is important, therefore, to extract the maximum amount of information from each photon.
So how many bits of information can realistically be extracted from a single photon? We can think of the fluorescent molecule as a transmitter sending signals to a receiver such as a photomultiplier tube. Under ideal conditions, transmitting one bit of information requires an energy approximately equal to the temperature of the environment (T) multiplied by the Boltzmann constant (k). A photon of green light (such as those emitted by GFPs) has an energy of 95kT at normal biological temperatures, and so could, in principle, yield about 95 bits of information.
This is a very generous limit, however, and is only valid if the information can be perfectly decoded and if the number of photons emitted is very small. It would also require us to exploit the quantum properties of the photon far better than is normally possible. A typical fluorescent molecule will emit about 104 photons before it photobleaches, so taking these limitations into account we could hope, at best, for a megabit of information from each fluorophore. This suggests two ways of obtaining more information from biological systems: designing more efficient microscope systems that make better use of the available photons; and increasing the number of photons available by, for instance, designing new types of fluorescent tags.
Breaking the limit
In recent years, researchers have made spectacular advances in the amount of information that they have been able to obtain from samples, thanks to the advent of techniques that can break through the diffraction limit. These techniques can dramatically improve resolution, but at the price of using more light on the sample, which could damage it.
One of the first practical suggestions for moving beyond the diffraction limit was made in 1928 by the Irish physicist Edward Synge, following discussions he had with Albert Einstein. Synge considered what would happen if an aperture much smaller in size than the wavelength of the light passing through it is placed so close to the surface of a sample that the gap between it and the surface is smaller than this wavelength. He concluded that the light passing through the aperture would not have sufficient distance to diffract before hitting the sample and passing back through the aperture: very fine structures could, therefore, be resolved.
Testing Synge’s idea experimentally, however, requires the ability to very precisely position the aperture above the surface and to maintain its position while scanning takes place, which was not possible with the technology available in the 1920s. Indeed, it was not until 1972 that Eric Ash and colleagues at University College London demonstrated the feasibility of this concept. Ash and Nichols used 3 cm microwaves, which, thanks to their relatively long wavelengths, relaxed the mechanical requirements considerably.
It was then another decade before Dieter Pohl at the University of Basel in Switzerland successfully applied the method at optical wavelengths, spawning the technique now known as near-field scanning optical microscopy (NSOM). NSOM has regularly achieved resolutions of about 25 nm, but maintaining the probe very close to the sample still presents a considerable technical challenge.
NSOM is only really suitable for imaging structures on the surface of cells, but it nevertheless holds much promise for biological imaging. In particular, the technique’s superb resolution makes it great for imaging microdomains in cell membranes, and it has been used to good effect for this application by Michael Edidin and co-workers at Johns Hopkins University in the US. Studying these microdomains, which are also known as membrane rafts, has been a spectacularly productive area of cell biology in recent years because many cellular signalling processes appear to be directed through them. Bacteria and certain viruses, including HIV, also gain entry into cells via membrane rafts, which are usually about 40–100 nm in diameter, and it appears that Alzheimer’s disease and some of the prion-related diseases (like nvCJD) are also associated with the properties of microdomains.
NSOM is a derivative of scanning probe techniques such as atomic force microscopy or scanning tunnelling microscopy rather than optical microscopy. A super-resolving technique based on far-field optical microscopy, on the other hand, would have several advantages over NSOM, such as allowing 3D imaging, reducing the imaging time and making the sample easier to manipulate. Several such techniques have emerged in recent years, all of which exploit some sort of nonlinear relationship between the excitation, or input signal, and the fluorescence, or output signal.
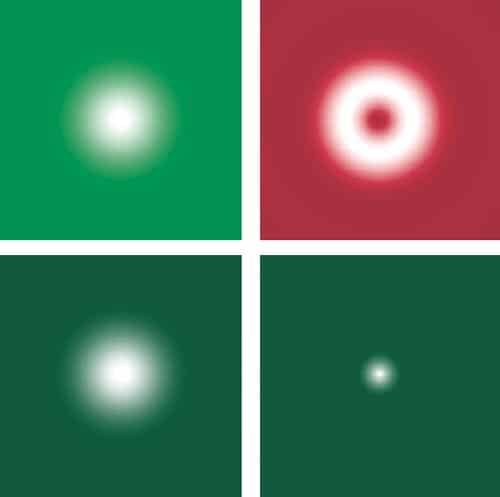
One of these techniques is stimulated emission and depletion (STED) microscopy, which has been developed over the last decade by Stefan Hell and coworkers at the Max Planck Institute for Biophysical Chemistry in Göttingen, Germany. STED is based on the idea that the resolution achievable with fluorescence microscopy can be improved by narrowing the effective width of the irradiation spot to below the diffraction limit so that the fluorescence used to build up the image emerges only from a small region. This is achieved by exciting fluorophores as normal with a diffraction-limited beam while a second beam — which has the same outer radius but is doughnut-shaped — de-excites the fluorophores in the outer part of the diffraction-limited spot via stimulated emission, thus preventing them from fluorescing (see “In good STED”).
Since the resulting fluorescence comes from an area much smaller than the diffraction limit, this technique can be used to achieve lateral resolutions down to about 30 nm, which is close to what can be achieved using NSOM. A STED module that can be used with a conventional scanning confocal microscope, which offers a lateral resolution of about 70 nm, is now available from the microscope manufacturer Leica, and Hell and co-workers reported last year in Science that this technique offers a valuable way of interrogating the microdomains in cell membranes.
A gathering STORM
Another fluorescence-microscopy method capable of excellent resolution is known as either photo-activated localization microscopy (PALM) or stochastic reconstruction microscopy (STORM). The first term is used by Eric Betzig, based at the Janelia Farm Research Campus in Virginia, US, who initiated the idea in 2006, while the latter term is used by Xiaowei Zhuang and colleagues at Harvard University, who have been actively developing the technique in the last couple of years. The method exploits the fact that single objects can be located with far greater precision than one can image two objects.
The task of locating the position of a fluorophore is essentially the same as locating the centre of the detected light distribution. If we detect a single photon from a fluorescent molecule, the position of the molecule can be typically located to within 200 nm or so, but this improves drastically if more photons can be detected. In one dimension, the spread of the light distribution shrinks by a factor of n as the number of detected photons increases by n2. In two dimensions, therefore, shrinking both dimensions by a factor of n requires n4 more photons. In other words, detecting 104 photons can reduce the radius of the patch to 20 nm.
STORM uses switchable fluorophores that can be rendered dark with one beam (usually from a red laser) and switched on with a second beam (which is usually green). During imaging, the fluorophores are first switched off by the red laser and then illuminated with the green laser so briefly that only a small proportion of the fluorophores in the field of view are switched on again. The result is that the distance between the active (fluorescing) molecules is greater than the diffraction limited resolution and they can therefore be located with great accuracy (see “A STORM cycle”).
A single cycle produces a sparse image made up of a few spots positioned very precisely. Each time the process is repeated, however, a different, random selection of molecules is switched on and so a similar sparse picture of points is recovered. By adding these sparse images together, a properly populated image is eventually built up. This technique can be applied to obtain a resolution of the order of 20 nm and can be used to generate 3D images. But since many imaging cycles are required, obtaining an image takes a long time and the sample is subject to a very high photon dose, which can harm live cells. Remarkable images of DNA molecules have been obtained with this method, but in its present incarnation it is not a prime candidate for live-cell imaging.
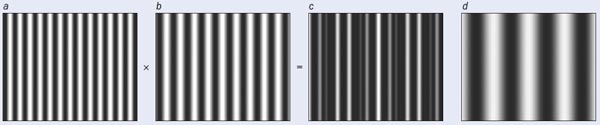
One problem with STED and STORM is that the equipment required for these techniques is much more complex than a conventional microscope. It is also possible, however, to achieve super-resolution with relatively small modifications to a standard full-field microscope, thanks to a technique suggested by Mats Gustafsson of the University of California at San Francisco in 2000 that is usually referred to as structured illumination microscopy. It exploits the fact that if a pattern (say produced by fluorophores in a sample) that is too fine to be imaged by a standard benchtop microscope is illuminated by light in a different pattern, then a set of low-resolution Moiré fringes is produced that are visible with the microscope. This pattern of fringes contains information about the original fine pattern. Once several Moiré images have been obtained, each with the illumination pattern at a different orientation to the sample, mathematical techniques can be used to reconstruct an image of the sample at enhanced resolution (see “Structured illumination microscopy”).
The degree to which the resolution is enhanced depends on the pattern used to illuminate the sample — using a sinusoidal grating pattern gives approximately twice the resolution of a conventional fluorescence microscope. If we increase the illumination intensity to the point where the fluorophores are driven into saturation (in other words, all of the illuminated fluorophores are raised to an excited state), however, we can obtain a lateral resolution of about 50 nm.
One advantage of structured illumination microscopy is that no scanning is required, which simplifies the optics. Another is that it utilizes the photons very efficiently, meaning that it is rather gentle on the sample compared with most other techniques for achieving super-resolution and so can be used for imaging live cells. In particular, the speed and convenience of structured illumination microscopy makes it ideal for high-resolution studies of dynamic processes on the cell membrane.
Lighting the way
These are just a few of the recent noteworthy advances in breaching the physical diffraction limits that have hindered measurements in cellular biology. What is fascinating is that the experimental needs of biology are driving developments in imaging technology, while advances in imaging technology are in turn inspiring new biological questions. Many of these developments are also going hand in hand with a revolution that is taking place in biological thinking, which intimately involves physicists. We are seeing a change in the nature of biological investigation as it takes on a sounder theoretical basis coupled to experimental analysis — the hallmarks of modern physics. These are exciting and interesting times to be working in biological research — and not just for biologists!
Box: Life changing physics
Developing imaging technologies for applications in cell biology has traditionally been hampered by formal academic divisions. For many years, mutual exchange between the essentially separate disciplines of physics and biology seemed impossible within the compartmentalized framework of professional 20th-century science. Of late, however, a new interdisciplinary zeal has gripped these two fields, drawing them together and blurring their traditional barriers. The reason for this is that biologists are now faced with challenges — such as imaging cellular function — that can only be addressed with theoretical and experimental tools that have in the past solely been used by physicists and engineers. The importance of a genuine interdisciplinary culture to deal with questions arising from the life sciences (as well as to generate new ones) has prompted the non-biological learned societies to promote such contact with their members. The Institute of Physics, for example, has recently established a Biological Physics Group with just such a purpose in mind. This group will have its inaugural showcase meeting, themed “Physics meets biology”, in Oxford this July.
Sceptical physicists might perhaps wonder how such contact benefits them — but biology in fact raises plenty of interesting physical issues. Single living cells grow, divide and respond to stimuli in a predictable manner, so their components (such as the cytoplasm and membrane rafts) must operate by defined sets of rules. These rules are out there waiting to be discovered. Similarly, when collections of cells work together (as in brain function) their collective or “emergent” behaviour can also, in principle, be predicted. Understanding these rules presents formidable challenges that will require biologists, physicists, mathematicians and engineers to all working together.
As well as these essentially theoretical challenges, cellular biology also presents physics with major measurement challenges. Biology is often said to be data rich, with large amounts of data to be processed, but given the awesome complexity of the problems being addressed, the subject is actually rather data poor. There are pressing needs, therefore, to develop better measurement techniques for studying both single living cells and dense cellular clusters. Cellular imaging offers one route that will contribute to these data-collection regimes and at the same time unites physicists, engineers and biologists on the path to common goals.
At a Glance: Super-resolution fluorescence microscopy
- Fluorescence microscopy, which uses optical microscopes to observe biological structures that have been tagged with fluorescent molecules, is a key tool used by biologists for studying cells
- Conventional fluorescence-microscopy systems are limited by the effects of diffraction, so the best resolution they can achieve is approximately 200 nm
- Many interesting and important cellular structures, however, are on length scales smaller than this, and in recent years biologists have turned to physicists for help in breaking through this diffraction limit
- The result is several novel techniques, including stimulated emission depletion microscopy (STED), stochastic reconstruction microscopy (STORM) and structured illumination microscopy, all of which are capable of resolving structures as small as 50 nm across
More about: Super-resolution fluorescence microscopy
M Gustafsson 2005 Nonlinear structured illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution Proc. Natl Acad. Sci. USA 102 13081
B Huang et al. 2008 Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy Science 319 810
T Klar et al. 2000 Fluorescence microscopy with diffraction resolution broken by stimulated emission Proc. Natl Acad. Sci. USA 97 8206
J J Sieber et al. 2007 Anatomy and dynamics of a supramolecular membrane protein cluster Science 317 1072
M Somekh et al. 2008 Resolution in structured illumination microscopy, a probabilistic approach J. Opt. Soc. Ameri. A at press
www.iop.org/activity/groups/subject/bp