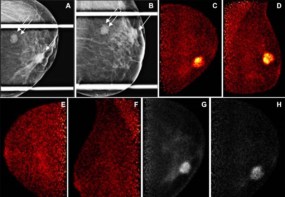
When it comes to diagnosing brain cancer, biopsies are often the first port of call. Surgeons begin by removing a thin layer of tissue from the tumour and examining it under a microscope, looking closely for signs of disease. However, not only are biopsies highly invasive, but the samples obtained only represent a fraction of the overall tumour site. MRI offers a less intrusive approach, but radiologists have to manually delineate the tumour area from the scan before they can classify it, which is time consuming.
Recently, scientists from the US developed a model capable of classifying numerous intracranial tumour types without the need for a scalpel. The model, called a convolutional neural network (CNN), uses deep learning – a type of machine learning algorithm found in image recognition software – to recognize these tumours in MR images, based on hierarchical features such as location and morphology. The team’s CNN could accurately classify several brain cancers with no manual interaction.
“This network is the first step toward developing an artificial intelligence-augmented radiology workflow that can support image interpretation by providing quantitative information and statistics,” says first author Satrajit Chakrabarty.
Predicting tumour type
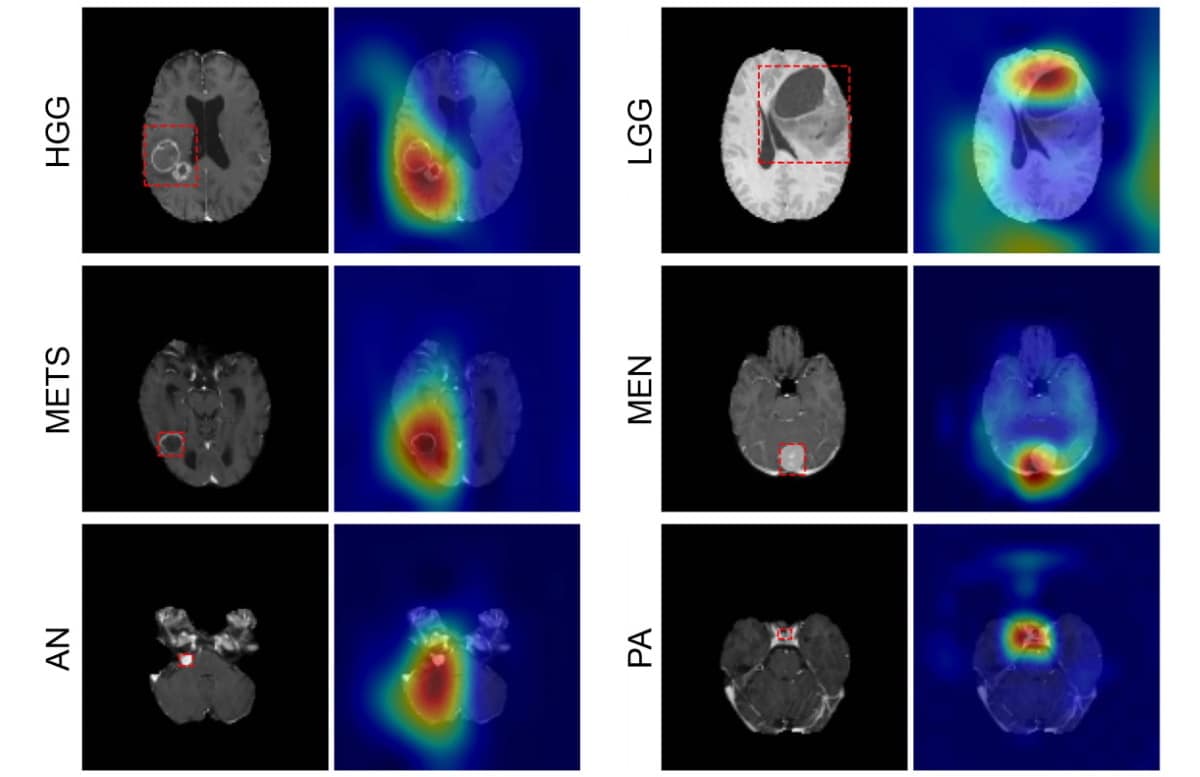
The CNN can detect six common types of intracranial tumour: high- and low-grade gliomas, meningioma, pituitary adenoma, acoustic neuroma and brain metastases. Writing in Radiology: Artificial Intelligence, the team – led by Aristeidis Sotiras and Daniel Marcus at Washington University School of Medicine (WUSM) – says that this neural network is the first to directly determine tumour class, as well as detect the absence of a tumour, from a 3D magnetic resonance volume.
To ascertain the accuracy of their CNN, the researchers created two multi-institutional datasets of pre-operative, post-contrast MRI scans from four publicly available databases, alongside data obtained at WUSM.
The first, the internal dataset, contained 1757 scans across seven imaging classes: the six tumour classes and one healthy class. Of these scans, 1396 were training data, which the team used to teach the CNN how to discriminate between each class. The remaining 361 were subsequently used to test the performance of the model (internal test data).

The CNN correctly identified tumour type with 93.35% accuracy, as confirmed by the radiology reports associated with each scan. What’s more, the probability that a patient actually had the specific cancer that the CNN detected (rather than being healthy or having any other type of tumour) was 85–100%.
Few false negatives were observed across all imaging classes; the probability that patients who tested negative for a given class did not have that disease (or were not healthy) was 98–100%.
Next, the researchers tested their model against a second external dataset containing only high- and low-grade gliomas. These scans were sourced separately to those in the internal dataset.
“As deep-learning models are very sensitive to data, it has become standard to validate their performance on an independent dataset, obtained from a completely different source, to see how well they generalize [react to unseen data],” explains Chakrabarty.
Diffusion MRI and machine learning models classify childhood brain tumours
The CNN demonstrated good generalization capability, scoring an accuracy of 91.95% on the external test data. The results suggest that the model could help clinicians diagnose patients with the six tumour types studied. However, the researchers note several limitations to their model, including misclassification of tumour type and grade due to poor image contrast. These shortfalls may be caused by inconsistencies in the imaging protocols used at the five institutions.
Looking to the future, the team hopes to train the CNN further by incorporating additional tumour types and imaging modalities.