SuperNova Green (SNG) is an effective new tool for the deactivation of proteins within cells. Developed by Yemima Dani Riani and the Nagai lab at Osaka University, the tool consists of a genetically encoded protein that reacts to light by generating damaging reactive oxygen species (ROS) in a small area around the molecule. These singlet and superoxide radicals damage neighbouring proteins enough that they become deactivated. Such a tool joins a growing arsenal of so called “optogenetic” actuators: genetically encoded light-activated proteins that can switch on or off spatial signalling pathways within cells, locally, on the nanoscale (BMC Biology 16 50).
Why destroy proteins with light?
Signalling pathways in cells depend on multitudes of molecular machines convening, accumulating and splitting apart again, in a vast molecular communication network. Super resolution microscopy has confirmed that there is a nanoscale component to this regulation that allows cells to elicit highly adaptive control over cell function. Direct links to such function have so far been hard to discern, and one reason is that the tools for altering such pathways on the nanoscale don’t exist. Dani Riani and colleagues have now introduced a new iteration of optogenetic tool, which can be hardwired into cells, fused to a protein and targeted for deactivation using laser light.
Depending on the size of the area illuminated with blue light, the length scale of protein deactivation can be altered. By concentrating the beam, single groups of proteins can be targeted, and by widening the beam, entire cells can be ablated, while their neighbours remain intact.

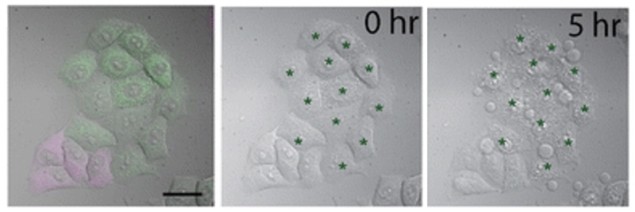
In the study, the Nagai group showcased whole-cell ablation. By selectively targeting the mitochondria in cancer cells, SNG could kill cells outright. The probe is relatively sensitive, so 2 min of illumination was all that was needed to kill the cells. This whole-cell application could be useful for studying the elasticity or mechanics of cells in the developing spinal cord (neural tube), for example, as an alternative to high powered laser ablation, which tends to damage nearby cells and makes measurement difficult.
The group showed that selective nanoscale protein inactivation is possible without significant damage to non-targeted proteins. By testing the effect of SNG on two tagged proteins that were very close to each other in the cell membrane, they demonstrated that only the SNG-tagged protein was affected. In this case, they targeted pleckstrin domains – short lengths of protein that bind to lipids in the cell membrane. When the pleckstrin domain is inactivated, the protein moves away from the membrane and into the cytoplasm. Sure enough, in these experiments, the signal in the cytoplasm only increased for SNG-tagged proteins, due to ROS-based destruction of the pleckstrin zone.
Why is SNG better?
Imaging multiple targets in cells allows for elements of signalling pathways to be visualized down a microscope. Much effort has gone into linking function to signalling, and one powerful method is to view both cause and effect at the same time. The authors point out that using SNG allows you to do this, by deactivating proteins with a short excitation wavelength (blue), while imaging something else, for example calcium dynamics, using a probe at a higher wavelength. This demonstrates a clear advantage of this new probe.
Additionally, SNG can deactivate proteins without flavin mononucleotide (FMN), which is present in many cancer cells, but is not always present in all somatic cells at the concentrations required by other optogenetic protein deactivators. SNG is also much simpler than other protein inactivation strategies, such as “anchoring away” (using a signal sequence to mislocalize a protein away from its functional location in a cell) or photo-induced oligomerization (engineering special proteins that bind to each other upon irradiation, and then fusing them to two separate proteins-of-interest).
Another method of deactivation is the use of the CRY2 system, which causes oligomerization of two protein targets, separately tagged with the two optogenetic components of the CRY2 system. The oligomerization causes non-physiological clusters, ablating protein function. The authors argue that their system is superior in its simplicity: only one laser is required for only one optogenetic protein, overcoming the barriers of cumbersome dual protein tagging, which can affect cell function and health.
Is SNG deactivation irreversible?
In the paper, the authors describe irreversible deactivation of a protein by SNG as an advantage for long-term study. This is of course true in theory – ROS-based damage to proteins affects covalent and ionic bonds, and is therefore irreparable. In practice, however, Dani Riani confirms that the protein has potential to be used for studying transient effects on very labile proteins such as cofilin – an actin regulator protein targeted by previous optogenetic tools developed in the Nagai lab.
Cytoskeletal regulation of nanocluster characteristics is still largely unexplored, as is the study of cell-wide changes resulting from the perturbation of a fast moving spatial signalling network. By using SNG to perturb such a protein (cofilin, for example), in conjunction with super resolution imaging of a related signalling protein (integrin, for example), researchers may have a powerful new tool to link nanoscale machinations to cell function.



