The conventional resolution limit for biological samples in atomic force microscopy (AFM) has be broken without needing to improve the physical experimental setup. George Heath and his team at Weill Cornell Medicine in New York City have done this by applying concepts of localization microscopy to molecular AFM studies and showed that AFM images can be given new life. The research, led by Simon Scheuring, is described in a recent paper in Nature.
Seeing molecular structure under the right conditions
Biomolecules are complex, dynamic structures that are difficult to study because they must be observed under conditions mimicking the environment of a real biological cell. Furthermore, sufficient resolution is needed in both space and time to fully capture molecular behaviour. AFM can already be used to image molecules at physiologically relevant temperature and pressure, and it can also be used in a high-speed mode (HS-AFM), where sufficient temporal resolution can be reached to observe conformational changes of molecules in real time.
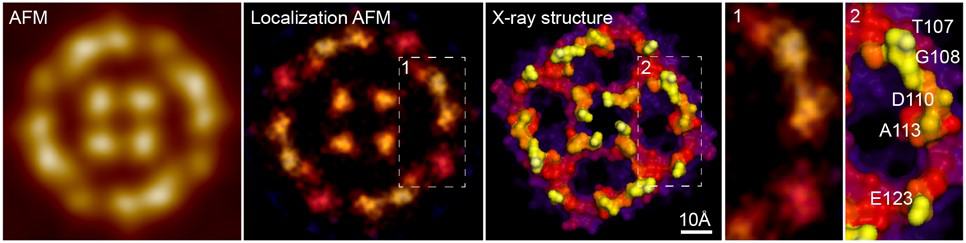
The remaining challenge had been obtaining sufficient spatial resolution at the sub-molecular scale. Until recently, AFM has been limited by the radii of AFM tips, the smallest of which are on the order of a few nanometres, and hence not sharp enough to resolve significant details on a single, corrugated, soft and flexible biomolecule in liquid. However, inspired by other super-resolution fluorescence microscopy techniques, Heath and his colleagues have now shown that resolution down to the angstrom-range (10-10 m) can be extracted from AFM images using a post-acquisition image reconstruction technique dubbed localization AFM (LAFM).
The impact of post-acquisition reconstruction
The LAFM approach is based on the idea that resolution superior to physical limitations can be achieved by determining the localization of isolated signals with high spatial precision using localization algorithms to process several images of the sample. The results are then merged to create a compiled map. Specifically for AFM, these signals are local maxima in the height traces – the locations where the force between the tip and the sample is largest. These are extracted from several images at specific locations and then merged in a peaking-probability map.
Scheuring describes the technique: “Imagine the high-resolution molecular features were marbles on a table, and you were trying to sense them by scanning a basketball over them. You most likely cannot resolve them. However, if you imagine that they, like individual ping-pong balls, stochastically bounce up, you will be able to localize them individually. Merging later all localization coordinates, you will be able to reconstruct the pattern of the marbles.”
New insight into biomolecular properties
Using LAFM, the team resolved single features in the surface protruding loops from aquaporin-Z (AqpZ) tetrameric channels. AqpZ is a protein that acts as a water channel in cell membranes. They were able to resolve details comparable to those in X-ray crystallographic structures, details that were previously undetectable in the AFM images (see figure). The team also produced videos showing pH-dependent conformational changes in the molecule CLC-ec1, which is found in many organisms and cells. This provided important information about the transport mechanism of this dimeric transporter molecule.

AFM detects heteroatoms in graphene nanoribbons
A LAFM map can be reconstructed either by recording many molecules in a few frames, or by observing a single molecule over a longer time. This makes it possible to observe either time- or environment-dependent conformational changes, but also acquire high-resolution information of individual molecules. Such capabilities can help to significantly advance understanding of biomolecular structure and function in the future, and Scheuring hopes that LAFM maps will become the new standard for biomolecular imaging.