Incorporating ventilation images into radiotherapy plans to treat lung cancer could reduce the incidence of debilitating radiation-induced lung injuries, such as radiation pneumonitis and radiation fibrosis. Specifically, ventilation imaging can be used to adapt radiation treatment plans to reduce the dose to high-functioning lung.
Positron emission tomography (PET) and single-photon emission computed tomography (SPECT) scans are the gold standard of ventilation imaging. However, these modalities are not always readily available and the cost of such exams may be prohibitive. As such, researchers are investigating the feasibility of alternatives such as MR or CT ventilation imaging.
CT ventilation imaging (CTVI) uses a treatment planning 4D-CT scan to estimate ventilation in the lungs. Conventional CTVIs rely on deformable image registration (DIR) of the inhalation and exhalation respiratory phases of a 4D-CT and the application of a ventilation metric to estimate ventilation. The key benefit of this approach is that CT images are typically available from exams performed for treatment planning, thus reducing the clinical time and the costs associated with nuclear medicine ventilation imaging.
Researchers at the University of Sydney recently investigated the use of machine learning as an alternative to DIR-based methods for producing CTVIs. They successfully generated CTVIs from breath-hold CT (BHCT) image pairs within 10 s, using a laptop computer and without the need for DIR or ventilation metrics. Their achievements, described in Medical Physics, produced performance measures comparable with conventional DIR-based methods.
Lead author James Grover of the ACRF Image X Institute and colleagues examined inhale and exhale BHCT image pairs and corresponding Galligas (Ga-68 aerosol) PET image sets for 15 lung cancer patients enrolled in a previous CTVI study. They selected Galligas PET as the reference imaging modality as it offers higher resolution and sensitivity than SPECT ventilation, thereby providing high-resolution images to train the deep-learning algorithm.
Grover and colleagues trained a 2D U-Net style convolutional neural network to produce axial CTVIs, which were then assembled to provide a 3D ventilation map of the patient’s lungs. The input training images consisted of exhalation, inhalation and average BHCT images. The neural network established relationships between these axial input BHCT images and axial labelled Galligas PET images. The team employed eightfold cross-validation to measure the robustness and increase the validity of the results attained by the neural network.
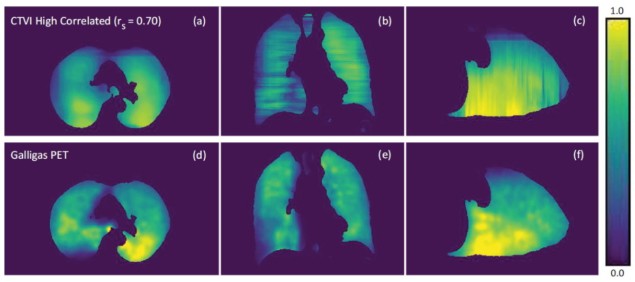
The researchers qualitatively assessed the neural network-produced CTVIs by visual comparison with the Galligas PET ventilation images. They report that the CTVIs tended to systematically overpredict ventilation within the lung when compared with the Galligas PET images. Each axial CTVI slice presented a smoothness among regions of low, medium and high ventilation, which caused difficulties in predicting small pockets of high and low ventilation within the lung. In the coronal and sagittal planes, ventilation maps showed distinct jagged edges in the superior–inferior direction.
For quantitative analysis, the team calculated the Spearman correlation and Dice similarity coefficient (DSC) between each patient’s CTVI and Galligas PET image. The DSC measured the spatial overlap between three equal lung sub-volumes, corresponding to high-, medium- and low-functioning lung, as defined by ventilation.
The mean Spearman correlation across the 15 patients was 0.58±0.14 (ranging from 0.28 to 0.70), while the mean DSCs over high-, medium- and low-functioning lung were 0.61±0.09, 0.43±0.05 and 0.62±0.07, respectively, with an average DSC of 0.55±0.06. The team note that these results are comparable to prior studies on CTVI generation.

Deep learning enables fast and accurate proton dose calculations
The researchers believe that the lower correlations seen for some patients are in part due to the use of a small patient dataset to train the neural network. They suggest that use of a 3D neural network would increase the Spearman correlation and DSC, as the model would be able to learn from a full patient volume instead of individual slices.
“We are planning to acquire patient ventilation images using a whole-body PET scanner to have the highest quality ground truth with which to develop the CTVI algorithms,” says Paul Keall, director of the ACRF Image X Institute. “We also hope to expand our investigations of CTVIs beyond lung cancer radiotherapy to use CTVI as a decision aid for surgical planning and early biomarker investigations across a range of respiratory diseases.”
![]() AI in Medical Physics Week is supported by Sun Nuclear, a manufacturer of patient safety solutions for radiation therapy and diagnostic imaging centres. Visit www.sunnuclear.com to find out more.
AI in Medical Physics Week is supported by Sun Nuclear, a manufacturer of patient safety solutions for radiation therapy and diagnostic imaging centres. Visit www.sunnuclear.com to find out more.